








生物技术通报 ›› 2025, Vol. 41 ›› Issue (9): 159-167.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0147
• 研究报告 • 上一篇
收稿日期:2025-02-13
出版日期:2025-09-26
发布日期:2025-09-24
通讯作者:
严倩,女,博士,副研究员,研究方向 :荔枝资源收集及评价;E-mail: yanqian@gdaas.cn作者简介:史发超,男,博士,助理研究员,研究方向 :荔枝遗传育种;E-mail: shifachao@126.com
基金资助:
SHI Fa-chao( ), JIANG Yong-hua, LIU Hai-lun, WEN Ying-jie, YAN Qian(
), JIANG Yong-hua, LIU Hai-lun, WEN Ying-jie, YAN Qian( )
)
Received:2025-02-13
Published:2025-09-26
Online:2025-09-24
摘要:
目的 TFL1基因是植物开花途径中的重要调控基因,克隆荔枝LcTFL1基因并构建系统发育树,分析其表达模式并进行功能研究,为荔枝成花调控提供靶标基因。 方法 基于前期获取的转录组数据,通过设计特异性引物,利用PCR技术从‘妃子笑’荔枝叶片中克隆得到LcTFL1基因,结合生物信息学分析其蛋白保守结构和系统进化关系,通过定量PCR技术选取荔枝不同组织,明确该基因的组织表达模式,利用亚细胞定位确定其蛋白表达位置,通过构建植物双源表达载体,异源过表达拟南芥,统计开花时间表型,分析验证LcTFL1基因功能。 结果 LcTFL1基因编码区全长为513 bp,编码170个氨基酸,为亲水性蛋白,具有保守PEBP家族结构域,启动子区域存在多个响应光、激素和胁迫的顺式作用元件,亚细胞定位显示其蛋白定位在细胞质中。系统进化关系表明荔枝LcTFL1基因和同科的文冠果(Xanthoceras sorbifolium)的JRO89相似性最高。组织表达分析显示,LcTFL1在种子的表达量最高,其次为顶芽和果实。在拟南芥中过表达LcTFL1基因,过表达拟南芥植株表现为始花期莲座叶片数量显著增多,叶片长度和宽度显著降低,且植株出现晚花表型。 结论 LcTFL1高表达会延迟拟南芥开花,作为调控开花的抑制因子存在。
史发超, 姜永华, 刘海伦, 文英杰, 严倩. 荔枝LcTFL1基因的克隆与功能分析[J]. 生物技术通报, 2025, 41(9): 159-167.
SHI Fa-chao, JIANG Yong-hua, LIU Hai-lun, WEN Ying-jie, YAN Qian. Cloning and Functional Analysis of LcTFL1 Gene in Litchi chinensis Sonn.[J]. Biotechnology Bulletin, 2025, 41(9): 159-167.
用途 Usage | 引物名称 Prime name | 序列 Sequence (5′-3′) |
|---|---|---|
克隆 Clone | LcTFL1-F | ATGACAGAAACACTATCT |
| LcTFL1-R | TCATCTTCTTCTAGCAGC | |
定位 Location | LcTFL1-F | CAGTGGTCTCACAACATGACAGAAACACTATCTGTGGGAAGAGTTG |
| LcTFL1-R | CAGTGGTCTCATACATCTTCTTCTAGCAGCGGTTTCTCTTTGG | |
过表达 Over-expression | LcTFL1-F | AACACGGGGGACTTTGCAACATGACAGAAACACTATC |
| LcTFL1-R | CCGTCGTGGTCTTTGTAATCTCTTCTTCTAGCAGCGG | |
内参 Actin GAPDH | Actin-F | ACCGTATGAGCAAGGAAATCACTG |
| Actin-R | TCGTCGTACTCACCCTTTGAAATC | |
荧光定量 RT-qPCR | LcTFL1-F | ATGGCCTCAGAGCTGCTTAT |
| LcTFL1-R | GTGGTGCCAGGAATGTTTGT |
表1 引物列表
Table 1 Primer list
用途 Usage | 引物名称 Prime name | 序列 Sequence (5′-3′) |
|---|---|---|
克隆 Clone | LcTFL1-F | ATGACAGAAACACTATCT |
| LcTFL1-R | TCATCTTCTTCTAGCAGC | |
定位 Location | LcTFL1-F | CAGTGGTCTCACAACATGACAGAAACACTATCTGTGGGAAGAGTTG |
| LcTFL1-R | CAGTGGTCTCATACATCTTCTTCTAGCAGCGGTTTCTCTTTGG | |
过表达 Over-expression | LcTFL1-F | AACACGGGGGACTTTGCAACATGACAGAAACACTATC |
| LcTFL1-R | CCGTCGTGGTCTTTGTAATCTCTTCTTCTAGCAGCGG | |
内参 Actin GAPDH | Actin-F | ACCGTATGAGCAAGGAAATCACTG |
| Actin-R | TCGTCGTACTCACCCTTTGAAATC | |
荧光定量 RT-qPCR | LcTFL1-F | ATGGCCTCAGAGCTGCTTAT |
| LcTFL1-R | GTGGTGCCAGGAATGTTTGT |
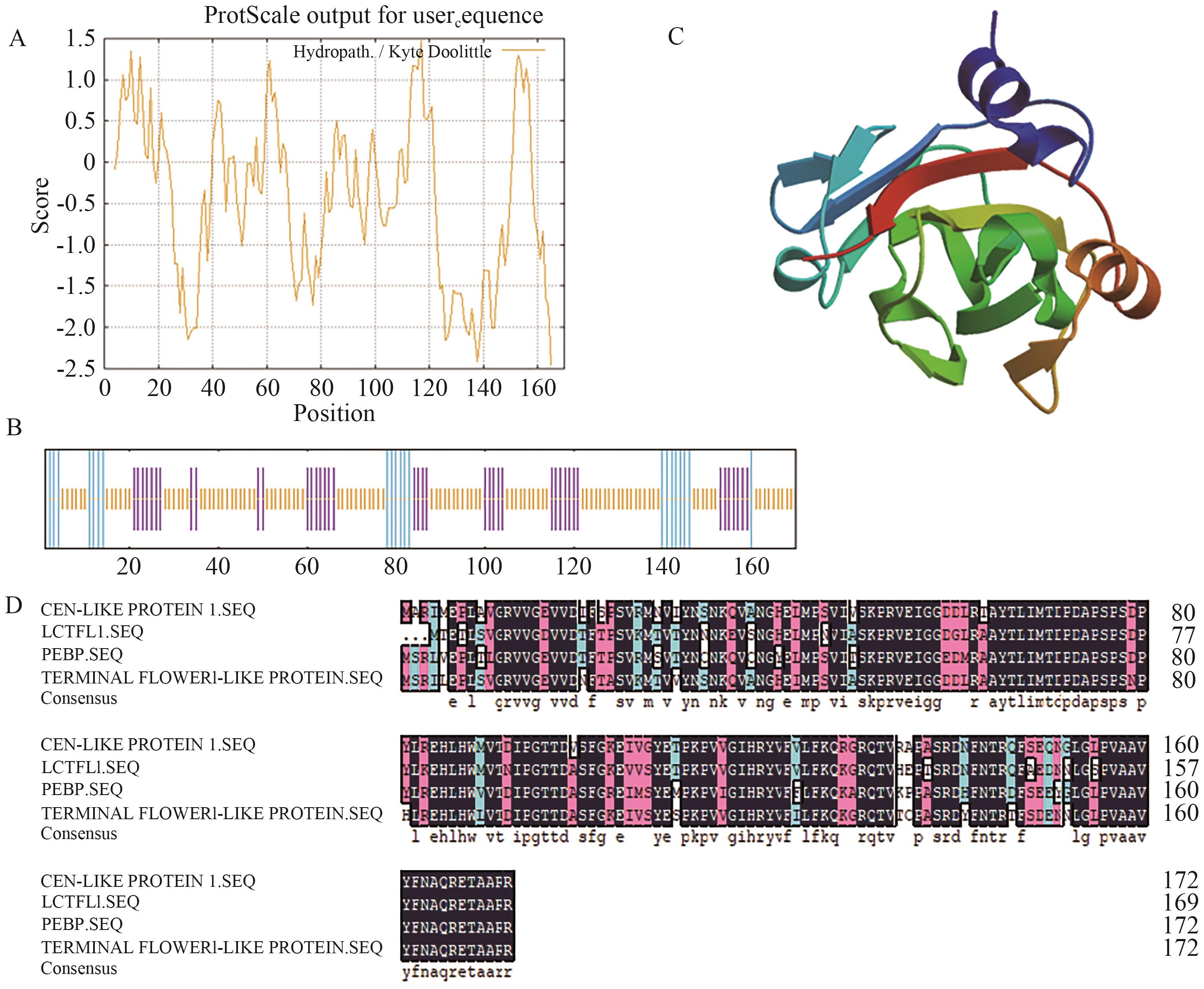
图1 LcTFL1蛋白生物信息学分析A:蛋白亲水/疏水性预测;B:蛋白二级结构预测;C:蛋白三级结构预测;D:蛋白序列比对
Fig. 1 Bioinformation analysis of LcTFL1A: Hydrophilic and hydrophobic prediction of protein. B: Secondary structure prediction of protein. C: Tertiary structure prediction of protein. D: Protein sequence alignment
| 元件类型 Type | 相关元件 Structure | 数量 Number | 序列 Sequence | 功能注释 Function annotation |
|---|---|---|---|---|
| 光响应元件 Light response element | TCT-motif | 1 | TCTTAC | Part of a light responsive element |
| TCCC-motif | 1 | TCTCCCT | Part of a light responsive element | |
| GATA-motif | 1 | AAGGATAAGG | Part of a light responsive element | |
| G-box | 1 | CACGTC | Part of a light responsive element | |
| G-Box | 1 | CACGTT | Part of a light responsive element | |
| Box 4 | 3 | ATTAAT | Part of a light responsive element | |
| ARE | 1 | AAACCA | Part of a light responsive element | |
| ACE | 1 | GACACGTATG | Part of a light responsive element | |
| circadian | 1 | CAAAGATATC | Involved in circadian control | |
| 激素响应元件 Hormone response element | ABRE | 2 | ACGTG | In the abscisic acid responsive |
| 胁迫响应元件Stress response element | TC-rich repeats | 2 | GTTTTCTTAC | Involved in defense and stress responsiveness |
| ARE | 1 | AAACCA | Anaerobic responsive element |
表2 LcTFL1启动子调控元件分析
Table 2 Regulatory element analysis of LcTFL1 promoter
| 元件类型 Type | 相关元件 Structure | 数量 Number | 序列 Sequence | 功能注释 Function annotation |
|---|---|---|---|---|
| 光响应元件 Light response element | TCT-motif | 1 | TCTTAC | Part of a light responsive element |
| TCCC-motif | 1 | TCTCCCT | Part of a light responsive element | |
| GATA-motif | 1 | AAGGATAAGG | Part of a light responsive element | |
| G-box | 1 | CACGTC | Part of a light responsive element | |
| G-Box | 1 | CACGTT | Part of a light responsive element | |
| Box 4 | 3 | ATTAAT | Part of a light responsive element | |
| ARE | 1 | AAACCA | Part of a light responsive element | |
| ACE | 1 | GACACGTATG | Part of a light responsive element | |
| circadian | 1 | CAAAGATATC | Involved in circadian control | |
| 激素响应元件 Hormone response element | ABRE | 2 | ACGTG | In the abscisic acid responsive |
| 胁迫响应元件Stress response element | TC-rich repeats | 2 | GTTTTCTTAC | Involved in defense and stress responsiveness |
| ARE | 1 | AAACCA | Anaerobic responsive element |
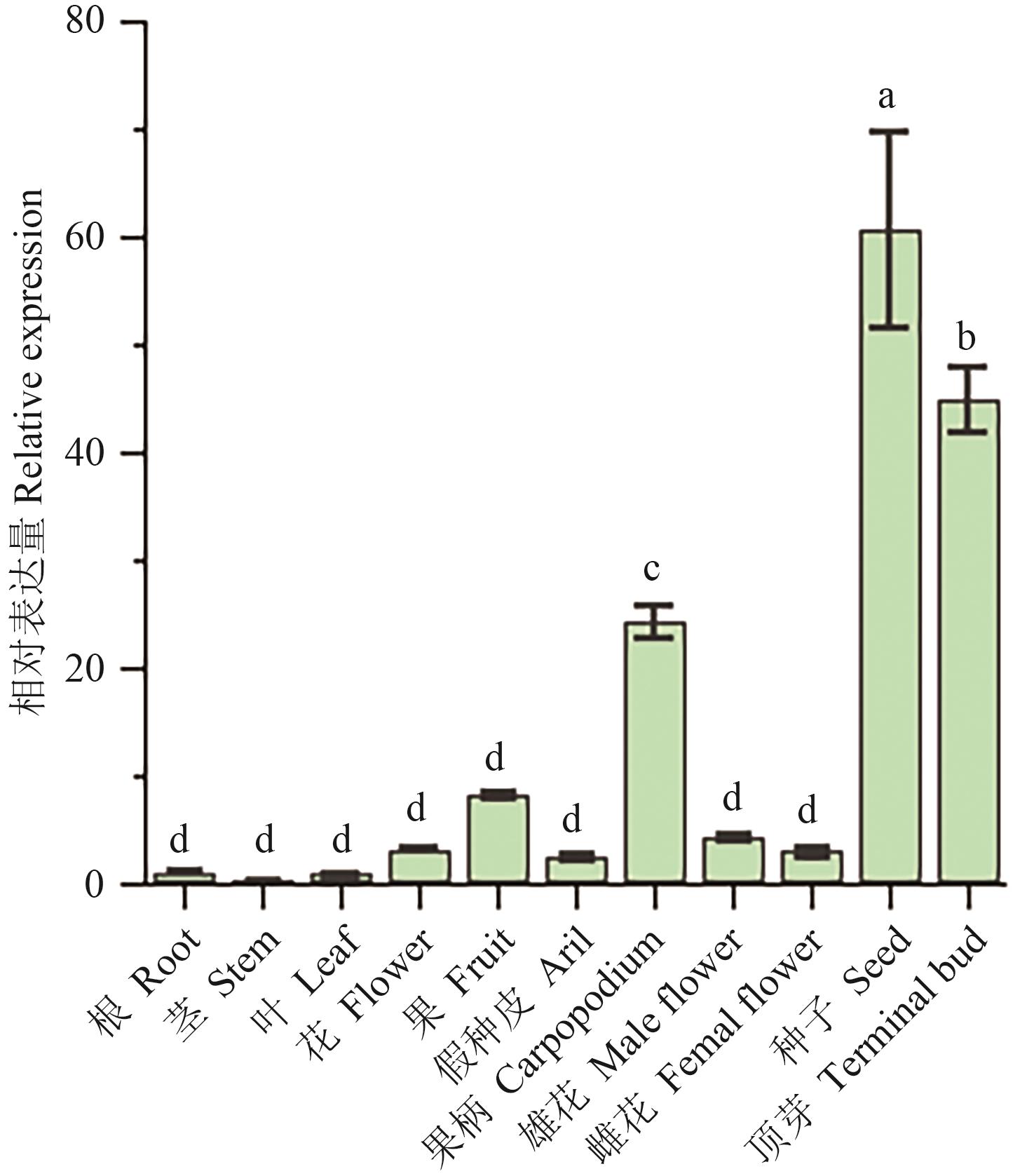
图3 荔枝LcTFL1不同组织表达分析不同字母表示差异显著(P<0.05)
Fig. 3 Expressions of LcTFL1 gene in different tissues in litchi (Litchi chinensis Sonn.)Different letters indicate significant difference (P<0.05)

图4 LcTFL1的亚细胞定位pBWA(V)HS-TFL1--GLosgfp在拟南芥原生质体中表达为绿色荧光蛋白,细胞核marker表达为红色荧光蛋白,紫色为叶绿体在特定波长下显示的紫色荧光
Fig. 4 Subcellular localization of LcTFL1The pBWA(V)HS-TFL1--GLosgfp is expressed as green fluorescent proteinin Arabidopsis thaliana protoplasts, the nuclear marker is expressed as red fluorescent protein, and purple is the purple fluorescence displayed by chloroplasts under specific wavelengths

图5 拟南芥转基因植株表型A、B:拟南芥野生型与过表达植株开花早晚形态;C:叶片数量及形态;D:叶片数量;E:叶片长度;F:叶片宽度。*代表差异显著(P<0.05),WT 代表野生型,OE(4-2)、OE(3-1)、OE(4-1)代表转基因株系
Fig. 5 Plant phenotype of transgenic ArabidopsisA, B: Phenotypic in tansgenic and wild-type Arabidopsis of flowering time. C: Number and morphology of leaves at bolting. D: Leaf number at bolting. E: Leaf length. F: Leaf width. * indicates significant difference at P<0.05. WT indicates wild type, OE (4-2), OE (3-1) and OE (4-1) indicate transgenic lines
| [1] | 李建国. 荔枝学 [M]. 北京: 中国农业出版社, 2008. |
| Li JG. The Litchi [M]. Beijing: China Agriculture Press, 2008. | |
| [2] | 文英杰, 欧良喜, 史发超, 等. 国家荔枝香蕉种质资源圃(广州)的荔枝资源保存现状及创新利用 [J]. 植物遗传资源学报, 2023, 24(5): 1205-1214. |
| Wen YJ, Ou LX, Shi FC, et al. Conservation status and innovative utilization of Litchi resources in the national Litchi and banana germplasm repository (Guangzhou) [J]. J Plant Genet Resour, 2023, 24(5): 1205-1214. | |
| [3] | Maple R, Zhu P, Hepworth J, et al. Flowering time: From physiology, through genetics to mechanism [J]. Plant Physiol, 2024, 195(1): 190-212. |
| [4] | Cao SH, Luo XM, Xu DA, et al. Genetic architecture underlying light and temperature mediated flowering in Arabidopsis, rice, and temperate cereals [J]. New Phytol, 2021, 230(5): 1731-1745. |
| [5] | Surkova SY, Samsonova MG. Mechanisms of vernalization-induced flowering in legumes [J]. Int J Mol Sci, 2022; 23(17):9889. |
| [6] | 齐仙惠, 巫东堂, 李改珍, 等. 拟南芥成花调控途径的研究进展 [J]. 山西农业大学学报: 自然科学版, 2018, 38(9): 1-7, 36. |
| Qi XH, Wu DT, Li GZ, et al. Regulation pathways of flowering in Arabidopsis thaliana [J]. J Shanxi Agric Univ Nat Sci Ed, 2018, 38(9): 1-7, 36. | |
| [7] | Zhu P, Lister C, Dean C. Cold-induced Arabidopsis FRIGIDA nuclear condensates for FLC repression [J]. Nature, 2021, 599(7886): 657-661. |
| [8] | Surkova SY, Samsonova MG. Mechanisms of vernalization-induced flowering in legumes [J]. Int J Mol Sci, 2022, 23(17): 9889. |
| [9] | Li ZC, He YH. Roles of brassinosteroids in plant reproduction [J]. Int J Mol Sci, 2020, 21(3): 872. |
| [10] | Gibbs DJ, Theodoulou FL, Bailey-Serres J. Primed to persevere: Hypoxia regulation from epigenome to protein accumulation in plants [J]. Plant Physiol, 2024, 197(1): kiae584. |
| [11] | Nielsen M, Menon G, Zhao YS, et al. COOLAIR and PRC2 function in parallel to silence FLC during vernalization [J]. Proc Natl Acad Sci USA, 2024, 121(4): e2311474121. |
| [12] | Zhang XP, Wang W, Zhu WD, et al. Mechanisms and functions of long non-coding RNAs at multiple regulatory levels [J]. Int J Mol Sci, 2019, 20(22): 5573. |
| [13] | Quiroz S, Yustis JC, Chávez-Hernández EC, et al. Beyond the genetic pathways, flowering regulation complexity in Arabidopsis thaliana [J]. Int J Mol Sci, 2021, 22(11): 5716. |
| [14] | Takagi H, Hempton AK, Imaizumi T. Photoperiodic flowering in Arabidopsis: Multilayered regulatory mechanisms of CONSTANS and the florigen FLOWERING LOCUS T [J]. Plant Commun, 2023, 4(3): 100552. |
| [15] | Shannon S, Meeks-Wagner DR. A mutation in the Arabidopsis TFL1 gene affects inflorescence meristem development [J]. Plant Cell, 1991, 3(9): 877-892. |
| [16] | Parcy F, Bomblies K, Weigel D. Interaction of LEAFY, AGAMOUS and TERMINAL FLOWER1 in maintaining floral meristem identity in Arabidopsis [J]. Development, 2002, 129(10): 2519-2527. |
| [17] | Lembinen S, Cieslak M, Zhang T, et al. Diversity of woodland strawberry inflorescences arises from heterochrony regulated by TERMINAL FLOWER 1 and FLOWERING LOCUS T [J]. Plant Cell, 2023, 35(6): 2079-2094. |
| [18] | Iwata H, Gaston A, Remay A, et al. The TFL1 homologue KSN is a regulator of continuous flowering in rose and strawberry [J]. Plant J, 2012, 69(1): 116-125. |
| [19] | Koskela EA, Mouhu K, Albani MC, et al. Mutation in TERMINAL FLOWER1 reverses the photoperiodic requirement for flowering in the wild strawberry Fragaria vesca [J]. Plant Physiol, 2012, 159(3): 1043-1054. |
| [20] | Mimida N, Kotoda N, Ueda T, et al. Four TFL1/CEN-like genes on distinct linkage groups show different expression patterns to regulate vegetative and reproductive development in apple (Malus × domestica Borkh.) [J]. Plant Cell Physiol, 2009, 50(2): 394-412. |
| [21] | Flachowsky H, Szankowski I, Waidmann S, et al. The MdTFL1 gene of apple (Malus × domestica Borkh.) reduces vegetative growth and generation time [J]. Tree Physiol, 2012, 32(10): 1288-1301. |
| [22] | Freiman A, Shlizerman L, Golobovitch S, et al. Development of a transgenic early flowering pear (Pyrus communis L.) genotype by RNAi silencing of PcTFL1-1 and PcTFL1-2 [J]. Planta, 2012, 235(6): 1239-1251. |
| [23] | Mohamed R, Wang CT, Ma C, et al. Populus CEN/TFL1 regulates first onset of flowering, axillary meristem identity and dormancy release in Populus [J]. Plant J, 2010, 62(4): 674-688. |
| [24] | Wang Z, Yang JC, Gao Q, et al. The transcription factor NtERF13a enhances abiotic stress tolerance and phenylpropanoid compounds biosynthesis in tobacco [J]. Plant Science, 2023, 334: 111772. |
| [25] | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔC T method [J]. Methods, 2001, 25(4): 402-408. |
| [26] | Serrazina S, Machado H, Costa RL, et al. Expression of Castanea crenata allene oxide synthase in Arabidopsis improves the defense to Phytophthora cinnamomi [J]. Front Plant Sci, 2021, 12: 628697. |
| [27] | Bellinazzo F, Nadal Bigas J, Hogers RAH, et al. Evolutionary origin and functional investigation of the widely conserved plant pebp gene stepmother of ft and tfl1 (smft) [J]. Plant J, 2024, 120(4): 1410-1420. |
| [28] | Lu XY, Lü PT, Liu H, et al. Identification of chilling accumulation-associated genes for Litchi flowering by transcriptome-based genome-wide association studies [J]. Front Plant Sci, 2022, 13: 819188. |
| [29] | Varkonyi-Gasic E, Moss SMA, Voogd C, et al. Homologs of FT, CEN and FD respond to developmental and environmental signals affecting growth and flowering in the perennial vine kiwifruit [J]. New Phytol, 2013, 198(3): 732-746. |
| [30] | 赵建文. 受激素和干旱调控的PhePEBP家族基因与毛竹笋芽萌发相关 [D]. 杭州: 浙江农林大学, 2019. |
| Zhao JW. PhePEBP family genes regulated by hormones and drought are related to bamboo shoot bud germination [D]. Hangzhou: Zhejiang A & F University, 2019. | |
| [31] | 王寻, 高凝, 张富军, 等. 苹果磷脂酰乙醇胺结合蛋白PEBP家族基因的鉴定与比较分析 [J]. 植物生理学报, 2021, 57(10): 1996-2010. |
| Wang X, Gao N, Zhang FJ, et al. Identification and comparative analysis of phosphatidyl ethanolamine binding protein (PEBP) family gene in apple [J]. Plant Physiol J, 2021, 57(10): 1996-2010. | |
| [32] | Kotoda N, Wada M. MdTFL1, a TFL1-like gene of apple, retards the transition from the vegetative to reproductive phase in transgenic Arabidopsis [J]. Plant Sci, 2005, 168(1): 95-104. |
| [33] | Wang YH, He XH, Yu HX, et al. Overexpression of four MiTFL1 genes from mango delays the flowering time in transgenic Arabidopsis [J]. BMC Plant Biol, 2021, 21(1): 407. |
| [34] | Esumi T, Kitamura Y, Hagihara C, et al. Identification of a TFL1 ortholog in Japanese apricot (Prunus mume Sieb. et Zucc.) [J]. Sci Hortic, 2010, 125(4): 608-616. |
| [35] | Zeng HY, Lu YT, Yang XM, et al. Ectopic expression of the BoTFL1-like gene of Bambusa oldhamii delays blossoming in Arabidopsis thaliana and rescues the tfl1 mutant phenotype [J]. Genet Mol Res, 2015, 14(3): 9306-9317. |
| [36] | Patil HB, Chaurasia AK, Azeez A, et al. Characterization of two TERMINAL FLOWER1 homologs PgTFL1 and PgCENa from pomegranate (Punica granatum L.) [J]. Tree Physiol, 2018, 38(5): 772-784. |
| [37] | Kotoda N, Iwanami H, Takahashi S, et al. Antisense expression of MdTFL1, a TFL1-like gene, reduces the juvenile phase in apple [J]. Jashs, 131(1): 74-81. |
| [38] | Imamura T, Nakatsuka T, Higuchi A, et al. The gentian orthologs of the FT/TFL1 gene family control floral initiation in Gentiana [J]. Plant Cell Physiol, 2011, 52(6): 1031-1041. |
| [39] | Boss PK, Sreekantan L, Thomas MR. A grapevine TFL1 homologue can delay flowering and alter floral development when overexpressed in heterologous species [J]. Funct Plant Biol, 2006, 33(1): 31-41. |
| [1] | 董向向, 缪百灵, 许贺娟, 陈娟娟, 李亮杰, 龚守富, 朱庆松. 森林草莓FveBBX20基因的生物信息学分析及开花调控功能[J]. 生物技术通报, 2025, 41(9): 115-123. |
| [2] | 刘佳丽, 宋经荣, 赵文宇, 张馨元, 赵子洋, 曹一博, 张凌云. 蓝莓R2R3-MYB基因鉴定及类黄酮调控基因表达分析[J]. 生物技术通报, 2025, 41(9): 124-138. |
| [3] | 黄诗宇, 田姗姗, 杨天为, 高曼熔, 张尚文. 赤苍藤WRI1基因家族的全基因组鉴定及表达模式分析[J]. 生物技术通报, 2025, 41(8): 242-254. |
| [4] | 李开杰, 吴瑶, 李丹丹. 红花CtbHLH128基因克隆及调控干旱胁迫应答功能研究[J]. 生物技术通报, 2025, 41(8): 234-241. |
| [5] | 赖诗雨, 梁巧兰, 魏列新, 牛二波, 陈应娥, 周鑫, 杨思正, 王博. NbJAZ3在苜蓿花叶病毒侵染本氏烟过程中的作用[J]. 生物技术通报, 2025, 41(8): 186-196. |
| [6] | 李雅琼, 格桑拉毛, 陈启迪, 杨宇环, 何花转, 赵耀飞. 异源过表达高粱SbSnRK2.1增强拟南芥对盐胁迫的抗性[J]. 生物技术通报, 2025, 41(8): 115-123. |
| [7] | 康琴, 汪霞, 谌明洋, 徐静天, 陈诗兰, 廖平杨, 许文志, 吴卫, 徐东北. 薄荷UV-B受体基因McUVR8的克隆与表达分析[J]. 生物技术通报, 2025, 41(8): 255-266. |
| [8] | 王芳, 乔帅, 宋伟, 崔鹏娟, 廖安忠, 谭文芳, 杨松涛. 甘薯IbNRT2基因家族全基因组鉴定和表达分析[J]. 生物技术通报, 2025, 41(7): 193-204. |
| [9] | 程珊, 王会伟, 陈晨, 朱雅婧, 李春鑫, 别海, 王树峰, 陈献功, 张向歌. 油莎豆MYB转录因子基因CeMYB154克隆及耐盐功能分析[J]. 生物技术通报, 2025, 41(6): 218-228. |
| [10] | 许慧珍, SHANTWANA Ghimire, RAJU Kharel, 岳云, 司怀军, 唐勋. 马铃薯SUMO E3连接酶基因家族分析及StSIZ1基因的克隆与表达模式分析[J]. 生物技术通报, 2025, 41(6): 119-129. |
| [11] | 杜量衡, 唐黄磊, 张治国. 控制水稻光响应基因ELM1的图位克隆[J]. 生物技术通报, 2025, 41(5): 82-89. |
| [12] | 彭绍智, 王登科, 张祥, 戴雄泽, 徐昊, 邹学校. 辣椒CaFD1基因克隆、表达特征及功能验证[J]. 生物技术通报, 2025, 41(5): 153-164. |
| [13] | 王天禧, 杨炳松, 潘荣君, 盖文贤, 梁美霞. 苹果PLATZ基因家族鉴定及MdPLATZ9基因功能研究[J]. 生物技术通报, 2025, 41(4): 176-187. |
| [14] | 宋佳怡, 苏芸丽, 郑兴艳, 夏文念, 杨冬梅, 胡慧贞. 金鱼草Expansin基因家族鉴定及其抗核盘菌相关基因筛选[J]. 生物技术通报, 2025, 41(4): 227-242. |
| [15] | 张益瑄, 马宇, 王童童, 盛苏奥, 宋家凤, 吕钊彦, 朱晓彪, 侯华兰. 马铃薯DIR家族全基因组鉴定及表达模式分析[J]. 生物技术通报, 2025, 41(3): 123-136. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||