








生物技术通报 ›› 2021, Vol. 37 ›› Issue (11): 190-196.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0235
杨华杰( ), 周玉萍, 范甜, 吕天晓, 谢楚萍, 田长恩(
), 周玉萍, 范甜, 吕天晓, 谢楚萍, 田长恩( )
)
收稿日期:2021-03-01
出版日期:2021-11-26
发布日期:2021-12-03
作者简介:杨华杰,男,硕士研究生,研究方向:分子遗传学;E-mail: 基金资助:
YANG Hua-jie( ), ZHOU Yu-ping, FAN Tian, LV Tian-xiao, XIE Chu-ping, TIAN Chang-en(
), ZHOU Yu-ping, FAN Tian, LV Tian-xiao, XIE Chu-ping, TIAN Chang-en( )
)
Received:2021-03-01
Published:2021-11-26
Online:2021-12-03
摘要:
拟南芥IQM4由AT2G26190编码,是1个含有IQ基序的Ca2+不依赖型钙调素结合蛋白,筛选和鉴定与IQM4互作的蛋白为进一步阐明IQM4所涉及的信号途径奠定基础。通过酵母双杂交筛选拟南芥幼苗cDNA文库,进而通过一对一的酵母双杂交试验和双分子荧光互补试验验证蛋白之间的互作关系。结果表明,获得15个与IQM4互作的候选蛋白;确定了CAT2和FBA1与IQM4蛋白存在互作关系,表明IQM4与CAT2及FBA1在植物细胞中存在相互作用。
杨华杰, 周玉萍, 范甜, 吕天晓, 谢楚萍, 田长恩. 拟南芥IQM4互作蛋白的筛选和鉴定[J]. 生物技术通报, 2021, 37(11): 190-196.
YANG Hua-jie, ZHOU Yu-ping, FAN Tian, LV Tian-xiao, XIE Chu-ping, TIAN Chang-en. Screening and Identification of IQM4-Interacting Proteins in Arabidopsis thaliana[J]. Biotechnology Bulletin, 2021, 37(11): 190-196.
| 基因Gene | 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|---|
| IQM4 | IQM4-F | TCAGAGGAGGACCTGCATATGATGGCTTCGGCTACTTTCTCTG |
| IQM4-R | CTAGTTATGCGGCCGCTGCAGCTAACAGTTTACTTGAACTCTAGGGCTT | |
| BF-IQM4—F | CGAGCTCAAGCTTCGAATTCGATGGGTCTTTCTCTTTCTTTGCTC | |
| BF-IQM4-75-R | GTACCGTCGACTGCAGAATTCCTAACAGTTTACTTGAACTCTAGGGCTT | |
| CAT2 | CAT2-F | GCCATGGAGGCCAGTGAATTCATGGATCCTTACAAGTATCGTCCAG |
| CAT2-R | ATGCCCACCCGGGTGGAATTCTTAGATGCTTGGTCTCACGTTCA | |
| BF-CAT2-F | TCGAGCTCAAGCTTCGAATTCGATGGATCCTTACAAGTATCGTCCAG | |
| BF-CAT2-R | GTACCGTCGACTGCAGAATTCTTAGATGCTTGGTCTCACGTTCA | |
| FBA1 | FBA1-F | GCCATGGAGGCCAGTGAATTCATGGCGTCAAGCACTGCG |
| FBA1-R | ATGCCCACCCGGGTGGAATTCTTAGTAGGTGTAGCCTTTTACAAACATAC | |
| BF-FBA1-F | TCGAGCTCAAGCTTCGAATTCGATGGCGTCAAGCACTGCG | |
| BF-FBA1-R | GTACCGTCGACTGCAGAATTCTTAGTAGGTGTAGCCTTTTACAAACATA |
表1 本试验所用的引物序列
Table 1 Primer sequences used in this experiment
| 基因Gene | 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|---|
| IQM4 | IQM4-F | TCAGAGGAGGACCTGCATATGATGGCTTCGGCTACTTTCTCTG |
| IQM4-R | CTAGTTATGCGGCCGCTGCAGCTAACAGTTTACTTGAACTCTAGGGCTT | |
| BF-IQM4—F | CGAGCTCAAGCTTCGAATTCGATGGGTCTTTCTCTTTCTTTGCTC | |
| BF-IQM4-75-R | GTACCGTCGACTGCAGAATTCCTAACAGTTTACTTGAACTCTAGGGCTT | |
| CAT2 | CAT2-F | GCCATGGAGGCCAGTGAATTCATGGATCCTTACAAGTATCGTCCAG |
| CAT2-R | ATGCCCACCCGGGTGGAATTCTTAGATGCTTGGTCTCACGTTCA | |
| BF-CAT2-F | TCGAGCTCAAGCTTCGAATTCGATGGATCCTTACAAGTATCGTCCAG | |
| BF-CAT2-R | GTACCGTCGACTGCAGAATTCTTAGATGCTTGGTCTCACGTTCA | |
| FBA1 | FBA1-F | GCCATGGAGGCCAGTGAATTCATGGCGTCAAGCACTGCG |
| FBA1-R | ATGCCCACCCGGGTGGAATTCTTAGTAGGTGTAGCCTTTTACAAACATAC | |
| BF-FBA1-F | TCGAGCTCAAGCTTCGAATTCGATGGCGTCAAGCACTGCG | |
| BF-FBA1-R | GTACCGTCGACTGCAGAATTCTTAGTAGGTGTAGCCTTTTACAAACATA |
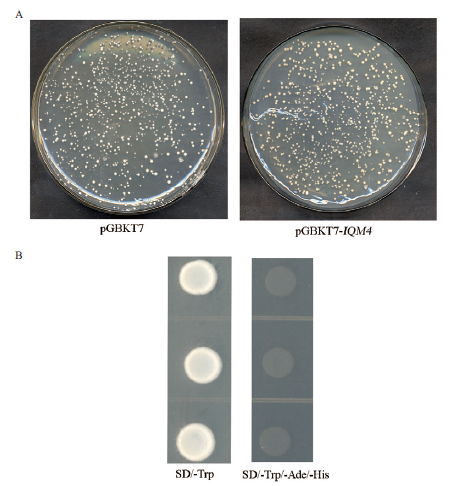
图2 IQM4蛋白自激活及毒性检测 A:pGBKT7-IQM4所表达的融合蛋白毒性的检测;B:IQM4自激活效应的检测
Fig. 2 Self-activation and toxicity detection of IQM4 protein A:Toxicity detection of the fusion protein expressed by pGBKT7-IQM4.B:Detec-tion of self-activation effect of IQM4
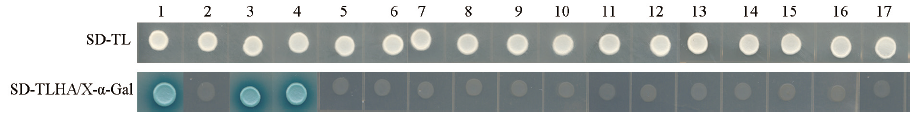
图4 IQM4互作候选蛋白的酵母双验证 SD-TL:缺Trp和Leu培养基;SD-TLHA/X-α-gal:缺Trp、Leu、His和Ade/+X-α-Gal培养基。1:pGADT7-T/pGBKT7-53,阳性对照;2:pGADT7/pGBKT7,阴性对照;3-17分别为BD-IQM4/AD-CAT2、FBA1、GAPA-2、ATDI19、FBA2、FBA5、LHCA2、LHCB1.1、LHB1B2、LHCB3、RBCS3B、CRB、CPN60B、RBCS1A和GAPC
Fig. 4 Validation of IQM4 interacting candidate proteins by yeast two hybrid system SD-TL:-Trp-Leu medium. SD-TLHA/X-α-Gal:-Trp-Leu-His-Ade /+X-α-Gal medium. 1:pGADT7-T/pGBKT7-53,positive control. 2:pGADT7/pGBKT7,negative control. 3-17 is BD-IQM4/AD-CAT2,FBA1,GAPA-2,ATDI19,FBA2,FBA5,LHCA2,LHCB1.1,LHB1B2,LHCB3,RBCS3B,CRB,CPN60B,RBCS1A,and GAPC,respectively
| [1] |
Poovaiah BW, Du LQ, Wang HZ, et al. Recent advances in calcium/calmodulin-mediated signaling with an emphasis on plant-microbe interactions[J]. Plant Physiol, 2013, 163(2): 531-542.
doi: 10.1104/pp.113.220780 pmid: 24014576 |
| [2] |
Perochon A, Aldon D, Galaud JP, et al. Calmodulin and calmodulin-like proteins in plant calcium signaling[J]. Biochimie, 2011, 93(12): 2048-2053.
doi: 10.1016/j.biochi.2011.07.012 URL |
| [3] | 曾后清, 张夏俊, 张亚仙, 等. 植物类钙调素生理功能的研究进展[J]. 中国科学:生命科学, 2016, 46(6): 705-715. |
|
Zeng HQ, Zhang XJ, Zhang YX, et al. Physiological functions of calmodulin-like proteins in plants[J]. Sci Sin:Vitae, 2016, 46(6): 705-715.
doi: 10.1360/N052015-00351 URL |
|
| [4] |
Dodd AN, Kudla J, Sanders D. The language of calcium signaling[J]. Annu Rev Plant Biol, 2010, 61: 593-620.
doi: 10.1146/arplant.2010.61.issue-1 URL |
| [5] | 曾后清, 张亚仙, 汪尚, 等. 植物钙/钙调素介导的信号转导系统[J]. 植物学报, 2016, 51(5): 705-723. |
| Zeng HQ, Zhang YX, Wang S, et al. Calcium/calmodulin-mediated signal transduction system in plants[J]. Chin Bull Bot, 2016, 51(5): 705-723. | |
| [6] |
Bouché N, Yellin A, Snedden WA, et al. Plant-specific calmodulin-binding proteins[J]. Annu Rev Plant Biol, 2005, 56(1): 435-466.
doi: 10.1146/arplant.2005.56.issue-1 URL |
| [7] |
Sanders D, Pelloux J, Brownlee C, et al. Calcium at the crossroads of signaling[J]. Plant Cell, 2002, 14(Suppl): S401-S417.
doi: 10.1105/tpc.002899 URL |
| [8] |
Lecourieux D, Ranjeva R, Pugin A. Calcium in plant defence-signalling pathways[J]. New Phytol, 2006, 171(2): 249-269.
pmid: 16866934 |
| [9] |
DeFalco TA, Bender KW, Snedden WA. Breaking the code:Ca2+ sensors in plant signalling[J]. Biochem J, 2010, 425(1): 27-40.
doi: 10.1042/BJ20091147 URL |
| [10] |
Bähler M, Rhoads A. Calmodulin signaling via the IQ motif[J]. FEBS Lett, 2002, 513(1): 107-113.
doi: 10.1016/S0014-5793(01)03239-2 URL |
| [11] | 田长恩, 周玉萍. 植物具IQ基序的钙调素结合蛋白的研究进展[J]. 植物学报, 2013, 48(4): 447-460. |
| Tian CE, Zhou YP. Research progress in plant IQ motif-containing calmodulin-binding proteins[J]. Chin Bull Bot, 2013, 48(4): 447-460. | |
| [12] |
Zhou YP, Chen YZ, Yamamoto KT, et al. Sequence and expression analysis of the Arabidopsis IQM family[J]. Acta Physiol Plant, 2010, 32(1): 191-198.
doi: 10.1007/s11738-009-0398-9 URL |
| [13] | 莫忠蓁, 黄章科, 周玉萍, 等. 拟南芥IQM1涉及主根生长对茉莉酸甲酯的反应[J]. 生物技术通讯, 2015, 26(3): 399-402. |
|
Mo ZZ, Huang ZK, Zhou YP, et al. IOM1 is involved in response of primary root growth to methyl jasmonate in Arabidopsis[J]. Lett Biotechnol, 2015, 26(3): 399-402.
doi: 10.1023/B:BILE.0000018258.58314.5c URL |
|
| [14] |
Lv T, Li X, Fan T, et al. The calmodulin-binding protein IQM1 interacts with CATALASE2 to affect pathogen defense[J]. Plant Physiol, 2019, 181(3): 1314-1327.
doi: 10.1104/pp.19.01060 URL |
| [15] | 程静之. 拟南芥IQM2参与自主途径成花调控的功能解析[D]. 广州:广州大学, 2016. |
| Cheng JZ. Functional analysis of IQM2 on the autonomous pathway flowering time regulation in Arabidopsis[D]. Guangzhou:Guangzhou University, 2016. | |
| [16] | 徐浩, 冯奕嘉, 范甜, 等. 拟南芥IQM3基因突变减少幼苗的侧根数量和增加主根长度[J]. 植物生理学报, 2019, 55(5): 629-634. |
| Xu H, Feng YJ, Fan T, et al. Disruption of IQM3 reduces the number of lateral roots and increases the length of primary root in Arabidopsis seedlings[J]. Plant Physiol J, 2019, 55(5): 629-634. | |
| [17] |
Gong LP, Cheng JZ, Zhou YP, et al. Disruption of IQM5 delays flowering possibly through modulating the juvenile-to-adult transition[J]. Acta Physiol Plant, 2016, 39(1): 1-10.
doi: 10.1007/s11738-016-2300-x URL |
| [18] | 冯奕嘉, 徐浩, 范甜, 等. 拟南芥IQM6突变推迟远轴面表皮毛的发生[J]. 植物生理学报, 2019, 55(6): 729-735. |
| Feng YJ, Xu H, Fan T, et al. IQM6 mutantion delays initiation of abaxial trichomes in Arabidopsis[J]. Plant Physiol J, 2019, 55(6): 729-735. | |
| [19] |
Zhou YP, Wu JH, Xiao WH, et al. Arabidopsis IQM4, a novel calmodulin-binding protein, is involved with seed dormancy and germination in Arabidopsis[J]. Front Plant Sci, 2018, 9: 721.
doi: 10.3389/fpls.2018.00721 URL |
| [20] | 萧文慧. 拟南芥IQM4基因功能的初步研究[D]. 广州:广州大学, 2016. |
| Xiao WH. Preliminary studuies on the function of Arabidopisis IQM4 gene[D]. Guangzhou:Guangzhou University, 2016. | |
| [21] | Soellick TR, Uhrig JF. Development of an optimized interaction-mating protocol for large-scale yeast two-hybrid analyses[J]. Genome Biol, 2001, 2(12): RESEARCH0052. |
| [22] |
Miller KE, Kim Y, Huh WK, et al. Bimolecular fluorescence complementation(BiFC)analysis:advances and recent applications for genome-wide interaction studies[J]. J Mol Biol, 2015, 427(11): 2039-2055.
doi: 10.1016/j.jmb.2015.03.005 URL |
| [23] | 刘云芬, 王薇薇, 祖艳侠, 等. 过氧化氢酶在植物抗逆中的研究进展[J]. 大麦与谷类科学, 2019, 36(1): 5-8. |
| Liu YF, Wang WW, Zu YX, et al. Research progress on the effects of catalase on plant stress tolerance[J]. Barley Cereal Sci, 2019, 36(1): 5-8. | |
| [24] |
Edel KH, Kudla J. Integration of calcium and ABA signaling[J]. Curr Opin Plant Biol, 2016, 33: 83-91.
doi: S1369-5266(16)30093-0 pmid: 27366827 |
| [25] |
Xue GP, McIntyre CL, Glassop D, et al. Use of expression analysis to dissect alterations in carbohydrate metabolism in wheat leaves during drought stress[J]. Plant Mol Biol, 2008, 67(3): 197-214.
doi: 10.1007/s11103-008-9311-y URL |
| [1] | 王子颖, 龙晨洁, 范兆宇, 张蕾. 利用酵母双杂交系统筛选水稻中与OsCRK5互作蛋白[J]. 生物技术通报, 2023, 39(9): 117-125. |
| [2] | 温晓蕾, 李建嫄, 李娜, 张娜, 杨文香. 小麦叶锈菌与小麦互作的酵母双杂交cDNA文库构建与应用[J]. 生物技术通报, 2023, 39(9): 136-146. |
| [3] | 李宇, 李素贞, 陈茹梅, 卢海强. 植物bHLH转录因子调控铁稳态的研究进展[J]. 生物技术通报, 2023, 39(7): 26-36. |
| [4] | 李帜奇, 袁月, 苗荣庆, 庞秋颖, 张爱琴. 盐胁迫盐芥和拟南芥褪黑素含量及合成相关基因表达模式分析[J]. 生物技术通报, 2023, 39(5): 142-151. |
| [5] | 侯筱媛, 车郑郑, 李姮静, 杜崇玉, 胥倩, 王群青. 大豆膜系统cDNA文库的构建及大豆疫霉效应子PsAvr3a互作蛋白的筛选[J]. 生物技术通报, 2023, 39(4): 268-276. |
| [6] | 崔吉洁, 蔡文波, 庄庆辉, 高爱平, 黄建峰, 陈亚辉, 宋志忠. 杧果Fe-S簇装配基因MiISU1的生物学功能[J]. 生物技术通报, 2023, 39(2): 139-146. |
| [7] | 鄢梦雨, 韦晓薇, 曹婧, 兰海燕. 异子蓬SabHLH169基因的克隆及抗旱功能分析[J]. 生物技术通报, 2023, 39(11): 328-339. |
| [8] | 阮航, 多浩源, 范文艳, 吕清晗, 姜述君, 朱生伟. AtERF49在拟南芥应答盐碱胁迫中的作用[J]. 生物技术通报, 2023, 39(1): 150-156. |
| [9] | 林蓉, 郑月萍, 徐雪珍, 李丹丹, 郑志富. 拟南芥ACOL8基因在乙烯合成与响应中的功能分析[J]. 生物技术通报, 2023, 39(1): 157-165. |
| [10] | 蔡佳, 梁振宇, 黄瑜, 鲁义善, 施钢, 简纪常. 利用酵母双杂交系统筛选与鉴定石斑鱼EcBAG3互作因子[J]. 生物技术通报, 2022, 38(8): 77-83. |
| [11] | 高聪, 萧楚健, 鲁帅, 王苏蓉, 袁卉华, 曹云英. 氧化石墨烯对拟南芥生长的促进作用[J]. 生物技术通报, 2022, 38(6): 120-128. |
| [12] | 徐红云, 张明意. GRAS转录因子AtSCL4负调控拟南芥应答渗透胁迫[J]. 生物技术通报, 2022, 38(6): 129-135. |
| [13] | 古盼, 齐学影, 李莉, 张曦, 单晓昳. AtRGS1胞吞动态调控G蛋白参与拟南芥生长发育和抗性反应[J]. 生物技术通报, 2022, 38(6): 34-42. |
| [14] | 周娟, 阎晋东, 李新梅, 刘雪晴, 赵强, 赵小英. 拟南芥F-box蛋白FKF1与转录因子FUL互作调控开花研究[J]. 生物技术通报, 2022, 38(3): 1-8. |
| [15] | 杨佳慧, 孙玉萍, 陆雅宁, 刘欢, 卢存福, 陈玉珍. 拟南芥AtTERT对大肠杆菌非生物胁迫抗性的影响[J]. 生物技术通报, 2022, 38(2): 1-9. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||