








生物技术通报 ›› 2022, Vol. 38 ›› Issue (3): 1-8.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0499
• 研究报告 • 下一篇
周娟( ), 阎晋东, 李新梅, 刘雪晴, 赵强, 赵小英(
), 阎晋东, 李新梅, 刘雪晴, 赵强, 赵小英( )
)
收稿日期:2021-04-16
出版日期:2022-03-26
发布日期:2022-04-06
作者简介:周娟,女,硕士研究生,研究方向:植物分子生物学;E-mail: 基金资助:
ZHOU Juan( ), YAN Jin-dong, LI Xin-mei, LIU Xue-qing, ZHAO Qiang, ZHAO Xiao-ying(
), YAN Jin-dong, LI Xin-mei, LIU Xue-qing, ZHAO Qiang, ZHAO Xiao-ying( )
)
Received:2021-04-16
Published:2022-03-26
Online:2022-04-06
摘要:
F-box蛋白FLAVIN-BINDING KELCH REPEAT F-BOX 1(FKF1)参与调控拟南芥光周期开花,但其分子机制尚不完全清楚。本研究通过体内和体外实验,证明FKF1与转录因子FRUITFULL(FUL)相互作用。qRT-PCR和Western blot结果显示,FKF1正调节FUL的转录水平,但不影响FUL蛋白的稳定性。遗传分析结果显示,35S-FKF1-Myc / ful-8双突变体的开花表型以及开花基因FLOWERING LOCUS T(FT)的转录水平与ful-8突变体相一致。研究结果表明FKF1可通过与FUL互作,在FUL的上游促进FT表达,进而促进开花。
周娟, 阎晋东, 李新梅, 刘雪晴, 赵强, 赵小英. 拟南芥F-box蛋白FKF1与转录因子FUL互作调控开花研究[J]. 生物技术通报, 2022, 38(3): 1-8.
ZHOU Juan, YAN Jin-dong, LI Xin-mei, LIU Xue-qing, ZHAO Qiang, ZHAO Xiao-ying. Study on the Interaction of F-box Protein FKF1 and Transcription Factor FUL in Regulating Flowering in Arabidopsis[J]. Biotechnology Bulletin, 2022, 38(3): 1-8.
| 基因座Locus | 蛋白名称Name of protein | 分值Score | 功能Function |
|---|---|---|---|
| AT1G22770 | Gigantea protein(GI) | 0.954 | Long day pathway |
| AT1G04400 | Cryptochrome 2(CRY2) | 0.952 | Light perception |
| AT5G60910 | AGAMOUS-like 8(FUL) | 0.946 | Floral promoter |
| AT3G42830 | RING/U-box superfamily protein(RBX1b) | 0.938 | |
| AT2G45660 | AGAMOUS-like 20(SOC1) | 0.9 | Floral promoter |
| AT5G20570 | RING-box 1(RBX1a) | 0.876 | |
| AT5G15840 | B-box type zinc finger protein with CCT domain(CO/BBX1) | 0.87 | Long day pathway |
| AT1G10940 | Protein kinase superfamily protein(SNRK1A) | 0.766 | |
| AT2G32950 | Transducin/WD40 repeat-like superfamily protein(COP1) | 0.726 | |
| AT1G09570 | Phytochrome A(PhyA) | 0.614 | Light perception |
| AT2G18790 | Phytochrome B(PhyB) | 0.57 | Light perception |
| AT5G11260 | Basic-leucine zipper(bZIP)transcription factor family protein(HY5) | 0.512 | |
| AT4G16250 | Phytochrome D(PhyD) | 0.87 | Light perception |
| AT2G25930 | Hydroxyproline-rich glycoprotein family protein(ELF3) | 0.878 | Circadian clock |
| AT2G46830 | Circadian clock associated 1(CCA1) | 0.608 | Circadian clock |
| AT1G09530 | Phytochrome interacting factor 3(PIF3) | 0.6 | Light signaling |
| AT5G61380 | CCT motif-containing response regulator protein(TOC1) | 0.888 | Circadian clock |
表1 FKF1候选互作蛋白
Table 1 Candidate interaction proteins of FKF1
| 基因座Locus | 蛋白名称Name of protein | 分值Score | 功能Function |
|---|---|---|---|
| AT1G22770 | Gigantea protein(GI) | 0.954 | Long day pathway |
| AT1G04400 | Cryptochrome 2(CRY2) | 0.952 | Light perception |
| AT5G60910 | AGAMOUS-like 8(FUL) | 0.946 | Floral promoter |
| AT3G42830 | RING/U-box superfamily protein(RBX1b) | 0.938 | |
| AT2G45660 | AGAMOUS-like 20(SOC1) | 0.9 | Floral promoter |
| AT5G20570 | RING-box 1(RBX1a) | 0.876 | |
| AT5G15840 | B-box type zinc finger protein with CCT domain(CO/BBX1) | 0.87 | Long day pathway |
| AT1G10940 | Protein kinase superfamily protein(SNRK1A) | 0.766 | |
| AT2G32950 | Transducin/WD40 repeat-like superfamily protein(COP1) | 0.726 | |
| AT1G09570 | Phytochrome A(PhyA) | 0.614 | Light perception |
| AT2G18790 | Phytochrome B(PhyB) | 0.57 | Light perception |
| AT5G11260 | Basic-leucine zipper(bZIP)transcription factor family protein(HY5) | 0.512 | |
| AT4G16250 | Phytochrome D(PhyD) | 0.87 | Light perception |
| AT2G25930 | Hydroxyproline-rich glycoprotein family protein(ELF3) | 0.878 | Circadian clock |
| AT2G46830 | Circadian clock associated 1(CCA1) | 0.608 | Circadian clock |
| AT1G09530 | Phytochrome interacting factor 3(PIF3) | 0.6 | Light signaling |
| AT5G61380 | CCT motif-containing response regulator protein(TOC1) | 0.888 | Circadian clock |
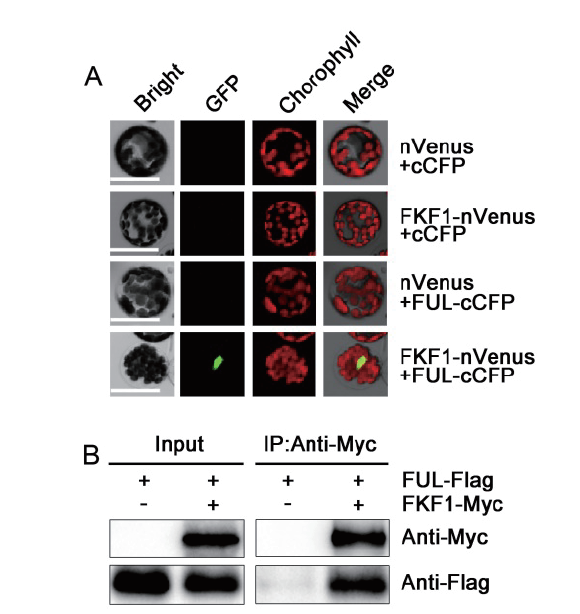
图2 双分子荧光互补(BiFC)和免疫共沉淀(Co-IP)实验分析FKF1与FUL的相互作用 A:BiFC 结果显示FKF1与FUL在拟南芥原生质体细胞核中相互作用。Bar = 50 μm;B:Co-IP结果显示FKF1与FUL存在相互作用
Fig. 2 BiFC and Co-IP assays showing FKF1 interaction with FUL A:BiFC assay showing the interaction of FKF1 with FUL in nucleus in Arabidopsis protoplast. Bar = 50μm. B:Co-IP assay showing the interaction of FKF1 with FUL
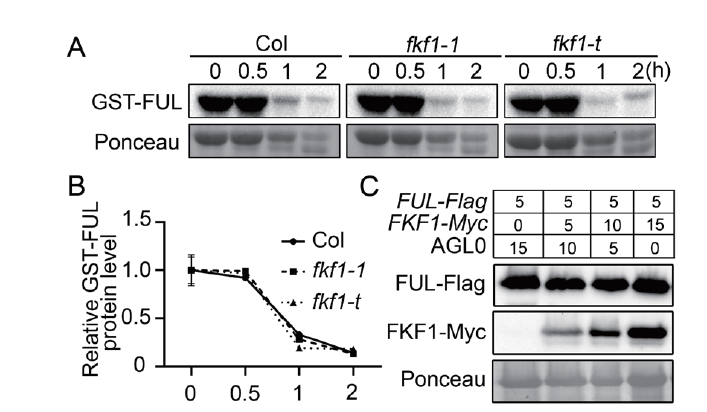
图3 FKF1对FUL蛋白稳定性的影响 A:FUL蛋白的半体内稳定性分析。将原核表达纯化得到的GST-FUL蛋白与野生型Col,fkf1-1或者fkf1-t 12 d苗龄幼苗的总蛋白等比例混合,孵育指定的时间,取样,用抗GST抗体进行免疫印迹检测。丽春红染色作为内标。进行了3次独立的实验,显示出相似的结果。B:(A)中GST-FUL的蛋白水平分析。利用丽春红染色信号将GST-FUL蛋白信号进行标准化。将开始取样时间点的值设定为1。误差棒代表着3个生物学重复标准偏差。C:FUL蛋白体内稳定性分析。将FUL-Flag和不同浓度FKF1-Myc在烟草叶片中共表达,提取总蛋白,用抗Flag和抗Myc抗体进行免疫印迹检测。数字表示共转化所用农杆菌的比例。丽春红染色作为内标。进行了3次独立的实验,显示出相似的结果
Fig. 3 Effect of FKF1 on FUL protein stability A:Semi-in vivo stability analysis of FUL protein. Escherichia coli-purified GST-FUL protein was mixed with total protein from the 12 d seedlings of wild-type Col,fkf1-1 or fkf1-t in equal proportion,and incubated for the given time. Sampled,and anti-GST antibody was detected by Western blotting. Ponceau staining was used as an internal standard. Three independent experiments were conducted,showing similar results. B:Protein level of GST-FUL in(A). GST-FUL protein level was normalized to Ponceau. The value of the starting point was set to 1. Bars refer to the standard deviations of three biological replicates. C:In vivo stability analysis of FUL protein. The FUL-Flag was co-expressed with increasing amounts of FKF1-Myc in tobacco leaves,and the total protein was extracted. The anti-Flag and anti-Myc proteins were detected by Western blotting,respectively. Numbers indicate the ratios of the concentrations of Agrobacteria used in co-infiltration. Ponceau staining was used as an internal standard. Three independent experiments were conducted,showing similar results
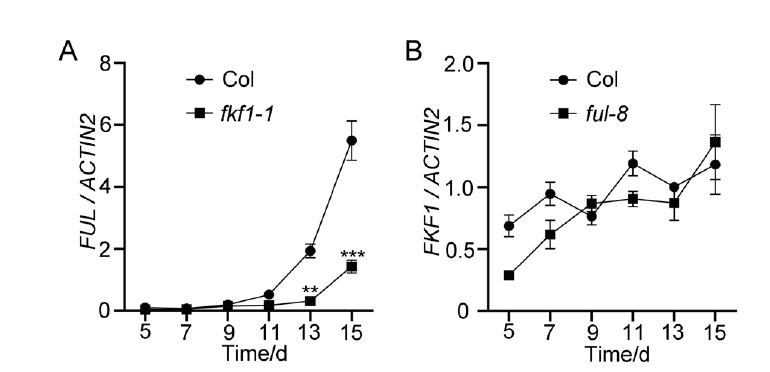
图4 FUL mRNA的表达受FKF1调节 A:野生型Col和fkf1-1植物中FUL mRNA表达分析;B:野生型Col和ful-8中FKF1 mRNA表达分析。将植物培养在1/2MS固体培养基上,长日照条件下生长不同天数,收集幼苗地上部分,用于qRT-PCR分析。ACTIN2作为内参基因。误差棒代表着3个生物学重复标准偏差。显著性差异:** P <0.01,*** P <0.001(Tukey检验用于检测统计学差异)
Fig. 4 FUL mRNA expression regulated by FKF1 A:mRNA expression of FUL in the wild-type Col and fkf1-1 mutant. B:mRNA expression of FKF1 in the wild-type Col and ful-8 mutant. Plants grew on 1/2 MS solid medium under long-day conditions(LDs),and aboveground seedlings were collected for quantitative real-time PCR(qRT-PCR)analysis. ACTIN2 served as the internal control. Error bars refer to the standard deviations of three biological replicates. Significant differences are indicated:**P<0.01,***P<0.001(Tukey's least significant difference test)
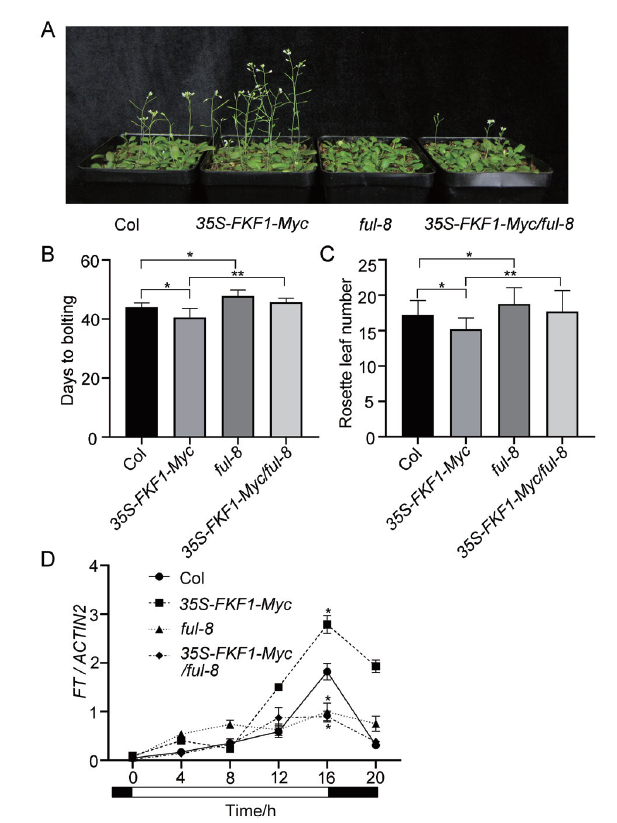
图5 Col,35S-FKF1-Myc,ful-8和35S-FKF1-Myc/ful-8植物开花表型 A:在长日照条件下生长40 d左右的植物照片;B:植物从播种到抽薹开花的时间;C:植物抽薹时的莲座叶数目;D:植物中FT的mRNA水平。将植物培养在1/2MS固体培养基上,长日照条件下生长,于第17天每4 h收集幼苗地上部分,用于qRT-PCR分析。ACTIN2作为内参基因。误差棒代表3个生物学重复的标准偏差。白色/黑色棒表示照光/黑暗相位。取样的第一个时间点作为0 h。误差棒代表标准偏差(n≥10)。显著性差异:* P <0.05,** P <0.01(Tukey检验用于检测统计学差异)
Fig. 5 Flowering phenotype of Col,35S-FKF1-Myc,ful-8 and 35S-FKF1-Myc / ful-8 plants A:Images of 40 d plants grown in soil under LDs. B:The days of plant sowed to bolting and blossoming of the respective genotypes. C:Number of rosette leaves at bolting of the respective genotypes. D:mRNA expression levels of FT in the respective genotypes. Plants grew in 1/2MS solid medium under LDs,samples were collected at every 4 h on day 17 for qRT-PCR analysis. ACTIN2 served as the internal control. Error bars refers to the standard deviations of three biological replicates. The white/black bars refer to light/dark phases. The time of collecting first sample is set as 0 h. Error bar refers to standard deviations(n≥10). Significant differences are indicated:*P<0.05 and **P<0.01(Tukey's least significant difference test)
| [1] |
Bernier G, Périlleux C. A physiological overview of the genetics of flowering time control[J]. Plant Biotechnol J, 2005, 3(1):3-16.
doi: 10.1111/pbi.2005.3.issue-1 URL |
| [2] | Fornara F, de Montaigu A, Coupland G. SnapShot:control of flowering in Arabidopsis[J]. Cell, 2010,141(3):550-550. e2. |
| [3] |
Srikanth A, Schmid M. Regulation of flowering time:all roads lead to Rome[J]. Cell Mol Life Sci, 2011, 68(12):2013-2037.
doi: 10.1007/s00018-011-0673-y pmid: 21611891 |
| [4] |
Wellmer F, Riechmann JL. Gene networks controlling the initiation of flower development[J]. Trends Genet, 2010, 26(12):519-527.
doi: 10.1016/j.tig.2010.09.001 pmid: 20947199 |
| [5] |
Kardailsky I, Shukla VK, Ahn JH, et al. Activation tagging of the floral inducer FT[J]. Science, 1999, 286(5446):1962-1965.
pmid: 10583961 |
| [6] |
Lee J, Oh M, Park H, et al. SOC1 translocated to the nucleus by interaction with AGL24 directly regulates leafy[J]. Plant J, 2008, 55(5):832-843.
doi: 10.1111/tpj.2008.55.issue-5 URL |
| [7] |
Zhang L, Chen L, Yu D. Transcription factor WRKY75 interacts with DELLA proteins to affect flowering[J]. Plant Physiol, 2018, 176(1):790-803.
doi: 10.1104/pp.17.00657 URL |
| [8] |
Lee H, Suh SS, Park E, et al. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis[J]. Genes Dev, 2000, 14(18):2366-2376.
doi: 10.1101/gad.813600 URL |
| [9] |
Yamaguchi A, Kobayashi Y, Goto K, et al. TWIN SISTER OF FT(TSF)Acts as a floral pathway integrator redundantly with FT[J]. Plant Cell Physiol, 2005, 46(8):1175-1189.
pmid: 15951566 |
| [10] |
Jang S, Torti S, Coupland G. Genetic and spatial interactions between FT, TSF and SVP during the early stages of floral induction in Arabidopsis[J]. Plant J, 2009, 60(4):614-625.
doi: 10.1111/tpj.2009.60.issue-4 URL |
| [11] |
Suárez-López P, Wheatley K, Robson F, et al. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis[J]. Nature, 2001, 410(6832):1116-1120.
doi: 10.1038/35074138 URL |
| [12] |
Valverde F. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering[J]. Science, 2004, 303(5660):1003-1006.
doi: 10.1126/science.1091761 URL |
| [13] |
Wigge PA. Integration of spatial and temporal information during floral induction in Arabidopsis[J]. Science, 2005, 309(5737):1056-1059.
doi: 10.1126/science.1114358 URL |
| [14] |
Imaizumi T, Tran HG, Swartz TE, et al. FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis[J]. Nature, 2003, 426(6964):302-306.
doi: 10.1038/nature02090 URL |
| [15] |
Nelson DC, Lasswell J, Rogg LE, et al. FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis[J]. Cell, 2000, 101(3):331-340.
pmid: 10847687 |
| [16] |
Imaizumi T, Schultz TF, Harmon FG, et al. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis[J]. Science, 2005, 309(5732):293-297.
pmid: 16002617 |
| [17] |
Sawa M, Nusinow DA, Kay SA, et al. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis[J]. Science, 2007, 318(5848):261-265.
doi: 10.1126/science.1146994 URL |
| [18] |
Song YH, Smith RW, To BJ, et al. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering[J]. Science, 2012, 336(6084):1045-1049.
doi: 10.1126/science.1219644 URL |
| [19] |
Lee BD, Kim MR, Kang MY, et al. The F-box protein FKF1 inhibits dimerization of COP1 in the control of photoperiodic flowering[J]. Nat Commun, 2017, 8(1):2259.
doi: 10.1038/s41467-017-02476-2 URL |
| [20] |
Lee BD, Cha JY, Kim MR, et al. Light-dependent suppression of COP1 multimeric complex formation is determined by the blue-light receptor FKF1 in Arabidopsis[J]. Biochem Biophys Res Commun, 2019, 508(1):191-197.
doi: 10.1016/j.bbrc.2018.11.032 URL |
| [21] |
Yan J, Li X, Zeng B, et al. FKF1 F-box protein promotes flowering in part by negatively regulating DELLA protein stability under long-day photoperiod in Arabidopsis[J]. J Integr Plant Biol, 2020, 62(11):1717-1740.
doi: 10.1111/jipb.v62.11 URL |
| [22] |
Wang JW, Czech B, Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana[J]. Cell, 2009, 138(4):738-749.
doi: 10.1016/j.cell.2009.06.014 URL |
| [23] |
Ferrándiz C, Gu Q, Martienssen R, et al. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER[J]. Development, 2000, 127(4):725-734.
pmid: 10648231 |
| [24] |
Melzer S, Lens F, Gennen J, et al. Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana[J]. Nat Genet, 2008, 40(12):1489-1492.
doi: 10.1038/ng.253 URL |
| [25] |
Mandel MA, Yanofsky MF. The Arabidopsis AGL8 MADS box gene is expressed in inflorescence meristems and is negatively regulated by APETALA1[J]. Plant Cell, 1995, 7(11):1763-1771.
pmid: 8535133 |
| [26] |
Gu Q, Ferrándiz C, Yanofsky MF, et al. The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development[J]. Development, 1998, 125(8):1509-1517.
pmid: 9502732 |
| [27] |
Schmid M, Uhlenhaut NH, Godard F, et al. Dissection of floral induction pathways using global expression analysis[J]. Development, 2003, 130(24):6001-6012.
doi: 10.1242/dev.00842 URL |
| [28] |
Abe M, Kobayashi Y, Yamamoto S, et al. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex[J]. Science, 2005, 309(5737):1052-1056.
doi: 10.1126/science.1115983 URL |
| [29] |
Torti S, Fornara F, Vincent C, et al. Analysis of the Arabidopsis shoot meristem transcriptome during floral transition identifies distinct regulatory patterns and a leucine-rich repeat protein that promotes flowering[J]. Plant Cell, 2012, 24(2):444-462.
doi: 10.1105/tpc.111.092791 URL |
| [30] |
Yamaguchi A, Wu MF, Yang L, et al. The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1[J]. Dev Cell, 2009, 17(2):268-278.
doi: 10.1016/j.devcel.2009.06.007 pmid: 19686687 |
| [31] |
Li W, Wang HP, Yu DQ. Arabidopsis WRKY transcription factors WRKY12 and WRKY13 oppositely regulate flowering under short-day conditions[J]. Mol Plant, 2016, 9(11):1492-1503.
doi: 10.1016/j.molp.2016.08.003 URL |
| [32] |
Hyun Y, Richter R, Vincent C, et al. Multi-layered regulation of SPL15 and cooperation with SOC1 integrate endogenous flowering pathways at the Arabidopsis shoot meristem[J]. Dev Cell, 2016, 37(3):254-266.
doi: 10.1016/j.devcel.2016.04.001 URL |
| [33] |
Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts:a versatile cell system for transient gene expression analysis[J]. Nat Protoc, 2007, 2(7):1565-1572.
doi: 10.1038/nprot.2007.199 URL |
| [34] |
Sparkes IA, Runions J, Kearns A, et al. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants[J]. Nat Protoc, 2006, 1(4):2019-2025.
pmid: 17487191 |
| [35] |
Moon J, Lee H, Kim M, et al. Analysis of flowering pathway integrators in Arabidopsis[J]. Plant Cell Physiol, 2005, 46(2):292-299.
doi: 10.1093/pcp/pci024 URL |
| [36] |
Oda A, Fujiwara S, Kamada H, et al. Antisense suppression of the Arabidopsis PIF3 gene does not affect circadian rhythms but causes early flowering and increases FT expression[J]. FEBS Lett, 2004, 557(1/2/3):259-264.
doi: 10.1016/S0014-5793(03)01470-4 URL |
| [37] |
Kim JJ, Lee JH, Kim W, et al. The microRNA156-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 module regulates ambient temperature-responsive flowering via FLOWERING LOCUS T in Arabidopsis[J]. Plant Physiol, 2012, 159(1):461-478.
doi: 10.1104/pp.111.192369 URL |
| [38] |
Lee JH, Yoo SJ, Park SH, et al. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis[J]. Genes Dev, 2007, 21(4):397-402.
doi: 10.1101/gad.1518407 URL |
| [39] |
Mathieu J, Yant LJ, Mürdter F, et al. Repression of flowering by the miR172 target SMZ[J]. PLoS Biol, 2009, 7(7):e1000148.
doi: 10.1371/journal.pbio.1000148 URL |
| [40] |
Searle I, He Y, Turck F, et al. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis[J]. Genes Dev, 2006, 20(7):898-912.
doi: 10.1101/gad.373506 URL |
| [41] |
Castillejo C, Pelaz S. The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering[J]. Curr Biol, 2008, 18(17):1338-1343.
doi: 10.1016/j.cub.2008.07.075 pmid: 18718758 |
| [42] |
Liu H, Yu X, Li K, et al. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis[J]. Science, 2008, 322(5907):1535-1539.
doi: 10.1126/science.1163927 URL |
| [43] |
Kumar SV, Lucyshyn D, Jaeger KE, et al. Transcription factor PIF4 controls the thermosensory activation of flowering[J]. Nature, 2012, 484(7393):242-245.
doi: 10.1038/nature10928 URL |
| [44] |
Aerts N, de Bruijn S, van Mourik H, et al. Comparative analysis of binding patterns of MADS-domain proteins in Arabidopsis thaliana[J]. BMC Plant Biol, 2018, 18(1):131.
doi: 10.1186/s12870-018-1348-8 URL |
| [45] |
Ito S, Song YH, Imaizumi T. LOV domain-containing F-box proteins:light-dependent protein degradation modules in Arabidopsis[J]. Mol Plant, 2012, 5(3):573-582.
doi: 10.1093/mp/sss013 URL |
| [1] | 王宝宝, 王海洋. 理想株型塑造之于玉米耐密改良[J]. 生物技术通报, 2023, 39(8): 11-30. |
| [2] | 李宇, 李素贞, 陈茹梅, 卢海强. 植物bHLH转录因子调控铁稳态的研究进展[J]. 生物技术通报, 2023, 39(7): 26-36. |
| [3] | 李帜奇, 袁月, 苗荣庆, 庞秋颖, 张爱琴. 盐胁迫盐芥和拟南芥褪黑素含量及合成相关基因表达模式分析[J]. 生物技术通报, 2023, 39(5): 142-151. |
| [4] | 崔吉洁, 蔡文波, 庄庆辉, 高爱平, 黄建峰, 陈亚辉, 宋志忠. 杧果Fe-S簇装配基因MiISU1的生物学功能[J]. 生物技术通报, 2023, 39(2): 139-146. |
| [5] | 鄢梦雨, 韦晓薇, 曹婧, 兰海燕. 异子蓬SabHLH169基因的克隆及抗旱功能分析[J]. 生物技术通报, 2023, 39(11): 328-339. |
| [6] | 阮航, 多浩源, 范文艳, 吕清晗, 姜述君, 朱生伟. AtERF49在拟南芥应答盐碱胁迫中的作用[J]. 生物技术通报, 2023, 39(1): 150-156. |
| [7] | 林蓉, 郑月萍, 徐雪珍, 李丹丹, 郑志富. 拟南芥ACOL8基因在乙烯合成与响应中的功能分析[J]. 生物技术通报, 2023, 39(1): 157-165. |
| [8] | 高聪, 萧楚健, 鲁帅, 王苏蓉, 袁卉华, 曹云英. 氧化石墨烯对拟南芥生长的促进作用[J]. 生物技术通报, 2022, 38(6): 120-128. |
| [9] | 徐红云, 张明意. GRAS转录因子AtSCL4负调控拟南芥应答渗透胁迫[J]. 生物技术通报, 2022, 38(6): 129-135. |
| [10] | 古盼, 齐学影, 李莉, 张曦, 单晓昳. AtRGS1胞吞动态调控G蛋白参与拟南芥生长发育和抗性反应[J]. 生物技术通报, 2022, 38(6): 34-42. |
| [11] | 杨佳慧, 孙玉萍, 陆雅宁, 刘欢, 卢存福, 陈玉珍. 拟南芥AtTERT对大肠杆菌非生物胁迫抗性的影响[J]. 生物技术通报, 2022, 38(2): 1-9. |
| [12] | 李兵娟, 郑璐, 沈仁芳, 兰平. 拟南芥RPP1A参与幼苗生长的蛋白质组学分析[J]. 生物技术通报, 2022, 38(2): 10-20. |
| [13] | 徐子涵, 刘倩, 苗大鹏, 陈跃, 胡凤荣. 春兰miR396过表达对拟南芥叶片生长、光合及叶绿素荧光特性的影响[J]. 生物技术通报, 2021, 37(5): 28-37. |
| [14] | 杨华杰, 周玉萍, 范甜, 吕天晓, 谢楚萍, 田长恩. 拟南芥IQM4互作蛋白的筛选和鉴定[J]. 生物技术通报, 2021, 37(11): 190-196. |
| [15] | 方丹丹, 张婷, 文晓鹏. 超表达马尾松PmPT3基因提高拟南芥耐低磷能力[J]. 生物技术通报, 2021, 37(10): 1-8. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||