








生物技术通报 ›› 2022, Vol. 38 ›› Issue (3): 9-15.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0802
收稿日期:2021-06-24
出版日期:2022-03-26
发布日期:2022-04-06
作者简介:张斌,男,博士,副教授,研究方向:植物发育生物学;E-mail: 基金资助:Received:2021-06-24
Published:2022-03-26
Online:2022-04-06
摘要:
盐胁迫是影响水稻生产的重要非生物胁迫之一。本研究以盐胁迫条件下的水稻转录组数据为材料,通过HTSeq和DESeq软件分析转录因子在转录组水平上的表达变化。主要结果如下:水稻在盐胁迫1、3和6 h三个时间点共有26个相同的差异表达转录因子,通过蛋白互作筛选出14个基因组成的重要模块;重要模块基因GO分析主要富集在ATP结合功能、对胁迫反应的过程以及细胞质膜囊泡组分上,KEGG分析主要富集在内质网蛋白质加工和内吞作用的通路上;同时鉴定出受盐胁迫诱导表达的关键热激转录因子基因Os03g0745000和Os06g0553100。推测这两个基因可能通过调控HSP70基因的转录表达,影响蛋白质折叠和组装,从而在水稻响应盐胁迫的过程中发挥作用。该研究为后续进行水稻遗传改良提供了候选基因,对进一步认识耐盐机制提供科学启示。
张斌, 杨昕霞. 水稻响应盐胁迫关键转录因子的鉴定[J]. 生物技术通报, 2022, 38(3): 9-15.
ZHANG Bin, YANG Xin-xia. Identification of Key Transcription Factors in Response to Salt Stress in Rice[J]. Biotechnology Bulletin, 2022, 38(3): 9-15.
| 基因IDGene ID | 正向引物Forward primer(5'-3') | 反向引物Reverse primer(5'-3') | 注释Description |
|---|---|---|---|
| Os03g0322900 | CCAAGAACAAGCTGGGCGAG | CTTGAACTCCGTCGCCTTCT | OsLEA3-2[ |
| Os11g0454300 | GCTCCAGCTCAAGCTCGTC | CATGGCATGCTGCTGCTC | RAB21[ |
| Os03g0745000 | CATTGCGCGTCATCGTGAAA | GACCTCGTACGTCTTGCACA | OsHsfA2a |
| Os06g0553100 | CCGTTCGTGTGGAAGACGTA | ACCATAGGTGTTGAGCTGGC | OsHsfC2b |
| Os10g0510000 | AGCTATCGTCCACAGGAA | ACCGGAGCTAATCAGAGT | OsActin |
| Os03g0277300 | ACCATCAAGAACGCCGTCAT | TGAGGACATTCTTCTCGCCG | OsHSP70 |
| Os03g0276500 | ATGATCGGCGTGCAGTACAA | GACGGTGCTTCCGAGATAGG | OsHSP71.1 |
表1 qRT-PCR反应引物序列和基因注释信息
Table 1 Primer sequence for qRT-PCR and gene annotation information
| 基因IDGene ID | 正向引物Forward primer(5'-3') | 反向引物Reverse primer(5'-3') | 注释Description |
|---|---|---|---|
| Os03g0322900 | CCAAGAACAAGCTGGGCGAG | CTTGAACTCCGTCGCCTTCT | OsLEA3-2[ |
| Os11g0454300 | GCTCCAGCTCAAGCTCGTC | CATGGCATGCTGCTGCTC | RAB21[ |
| Os03g0745000 | CATTGCGCGTCATCGTGAAA | GACCTCGTACGTCTTGCACA | OsHsfA2a |
| Os06g0553100 | CCGTTCGTGTGGAAGACGTA | ACCATAGGTGTTGAGCTGGC | OsHsfC2b |
| Os10g0510000 | AGCTATCGTCCACAGGAA | ACCGGAGCTAATCAGAGT | OsActin |
| Os03g0277300 | ACCATCAAGAACGCCGTCAT | TGAGGACATTCTTCTCGCCG | OsHSP70 |
| Os03g0276500 | ATGATCGGCGTGCAGTACAA | GACGGTGCTTCCGAGATAGG | OsHSP71.1 |
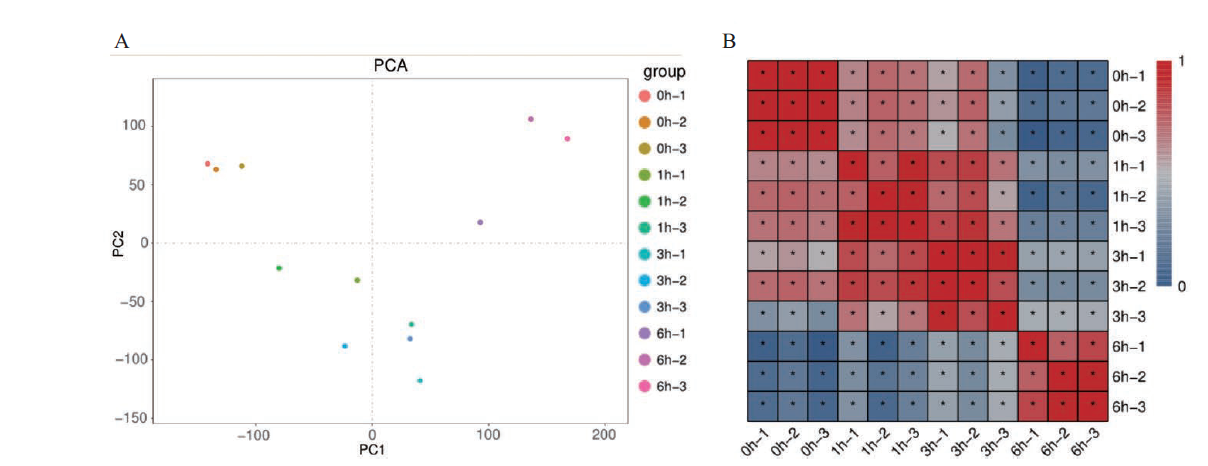
图1 测序样品关系 A:样品间PCA分析图;B:样品相关性热图。0 h、1 h、3 h和6 h表示0 h、1 h、3 h和6 h取样时间点,-1、-2和-3表示3个重复
Fig. 1 Relationship among sequencing samples A:PCA analysis diagram of samples. B:Heat map of sample correlation. 0,1,3 and 6 h represent 0,1,3 and 6 h sampling time points. -1,-2 and - 3 represent three repetitions

图2 盐胁迫差异表达基因和相同差异表达基因筛选图 A:盐胁迫1 h差异表达基因火山图;B:盐胁迫3 h差异表达基因火山图;C:盐胁迫6 h差异表达基因火山图;D:响应盐胁迫差异表达基因韦恩图
Fig. 2 Screening of differentially expressed genes(DEGs) and identifical DEGs under salt stress A:Differentially expressed genes under salt stress for 1 h. B:Volcano diagram of differentially expressed genes under salt stress for 3 h. C:Volcano diagram of differentially expressed genes under salt stress for 6 h. D:Venn diagram of differentially expressed genes in response to salt stress
| TF基因TF Gene | STRING中名称STRING ID | 注释Annotation |
|---|---|---|
| Os11g0151002 | OS11T0151002-00 | Os11g0151002蛋白质 Os11g0151002 protein |
| Os06g0553100 | OS06T0553100-01 | 热应激转录因子C-2b Heat stress transcription factor C-2b |
| Os04g0398000 | OS04T0398000-02 | Os04g0398000蛋白 Os04g0398000 protein |
| Os03g0820300 | OsJ_13136 | C2H2转录因子 C2H2 transcription factor |
| Os09g0522100 | DREB1H | 介导高盐和脱水诱导的转录 Mediates high salinity- and dehydration-inducible transcription |
| Os07g0688200 | OS07T0688200-01 | MYB转录因子 MYB transcription factor |
| Os03g0745000 | Hsf4 | 热应激转录因子A-2a Heat stress transcription factor A-2a |
| Os08g0200600 | OS08T0200600-01 | Os08g0200600蛋白质 Os08g0200600 protein |
| Os05g0437100 | OS05T0437100-00 | Os05g0437133蛋白质 Os05g0437133 protein |
| Os12g0564100 | OS12T0564100-01 | 无注释 Annotation not available |
| Os06g0670300 | P0485A07.2 | Os06g0670300蛋白 Os06g0670300 protein |
| Os02g0638650 | P0010C01.6 | 含AP2结构域的类转录因子 AP2 domain-containing transcription factor-like |
| Os01g0693400 | OS01T0693400-01 | 含AP2/ERF和B3结构域的蛋白质 AP2/ERF and B3 domain-containing protein |
| Os11g0168500 | OS11T0168500-00 | 无注释 Annotation not available |
| Os01g0859100 | OS01T0859100-01 | Os01g0859100蛋白 Os01g0859100 protein |
| Os01g0318400 | OS01T0318400-00 | Os01g0318400蛋白质 Os01g0318400 protein |
| Os09g0522200 | ERF24 | 脱水反应元件结合蛋白1A Dehydration-responsive element-binding protein 1A |
| Os03g0820400 | ZFP37 | C2H2型锌指转录因子ZFP37 C2H2 type zinc finger transcription factor ZFP37 |
| Os10g0571600 | OS10T0571600-01 | Os10g0571600蛋白质 Os10g0571600 protein |
| Os01g0626400 | WRKY11 | WRKY转录因子11 WRKY transcription factor 11 |
| Os03g0838800 | OS03T0838800-00 | 锌指蛋白 Zinc finger protein |
| Os05g0132700 | OS05T0132700-01 | Os05g0132700蛋白质 Os05g0132700 protein |
| Os03g0287400 | OsJ_10424 | LOB结构域蛋白1 LOB domain protein 1 |
| Os04g0529100 | OS04T0529100-01 | Os04g0529100蛋白质 Os04g0529100 protein |
| Os11g0154900 | OS11T0154900-01 | BZIP转录因子家族蛋白 BZIP transcription factor family protein |
| Os01g0298400 | OsJ_01424 | P型r2r3MYB蛋白 P-type r2r3 MYB protein |
表2 26个相同差异表达TF基因在STRING中的注释
Table 2 Notes of 26 identical differentially expressed TF genes in STRING
| TF基因TF Gene | STRING中名称STRING ID | 注释Annotation |
|---|---|---|
| Os11g0151002 | OS11T0151002-00 | Os11g0151002蛋白质 Os11g0151002 protein |
| Os06g0553100 | OS06T0553100-01 | 热应激转录因子C-2b Heat stress transcription factor C-2b |
| Os04g0398000 | OS04T0398000-02 | Os04g0398000蛋白 Os04g0398000 protein |
| Os03g0820300 | OsJ_13136 | C2H2转录因子 C2H2 transcription factor |
| Os09g0522100 | DREB1H | 介导高盐和脱水诱导的转录 Mediates high salinity- and dehydration-inducible transcription |
| Os07g0688200 | OS07T0688200-01 | MYB转录因子 MYB transcription factor |
| Os03g0745000 | Hsf4 | 热应激转录因子A-2a Heat stress transcription factor A-2a |
| Os08g0200600 | OS08T0200600-01 | Os08g0200600蛋白质 Os08g0200600 protein |
| Os05g0437100 | OS05T0437100-00 | Os05g0437133蛋白质 Os05g0437133 protein |
| Os12g0564100 | OS12T0564100-01 | 无注释 Annotation not available |
| Os06g0670300 | P0485A07.2 | Os06g0670300蛋白 Os06g0670300 protein |
| Os02g0638650 | P0010C01.6 | 含AP2结构域的类转录因子 AP2 domain-containing transcription factor-like |
| Os01g0693400 | OS01T0693400-01 | 含AP2/ERF和B3结构域的蛋白质 AP2/ERF and B3 domain-containing protein |
| Os11g0168500 | OS11T0168500-00 | 无注释 Annotation not available |
| Os01g0859100 | OS01T0859100-01 | Os01g0859100蛋白 Os01g0859100 protein |
| Os01g0318400 | OS01T0318400-00 | Os01g0318400蛋白质 Os01g0318400 protein |
| Os09g0522200 | ERF24 | 脱水反应元件结合蛋白1A Dehydration-responsive element-binding protein 1A |
| Os03g0820400 | ZFP37 | C2H2型锌指转录因子ZFP37 C2H2 type zinc finger transcription factor ZFP37 |
| Os10g0571600 | OS10T0571600-01 | Os10g0571600蛋白质 Os10g0571600 protein |
| Os01g0626400 | WRKY11 | WRKY转录因子11 WRKY transcription factor 11 |
| Os03g0838800 | OS03T0838800-00 | 锌指蛋白 Zinc finger protein |
| Os05g0132700 | OS05T0132700-01 | Os05g0132700蛋白质 Os05g0132700 protein |
| Os03g0287400 | OsJ_10424 | LOB结构域蛋白1 LOB domain protein 1 |
| Os04g0529100 | OS04T0529100-01 | Os04g0529100蛋白质 Os04g0529100 protein |
| Os11g0154900 | OS11T0154900-01 | BZIP转录因子家族蛋白 BZIP transcription factor family protein |
| Os01g0298400 | OsJ_01424 | P型r2r3MYB蛋白 P-type r2r3 MYB protein |
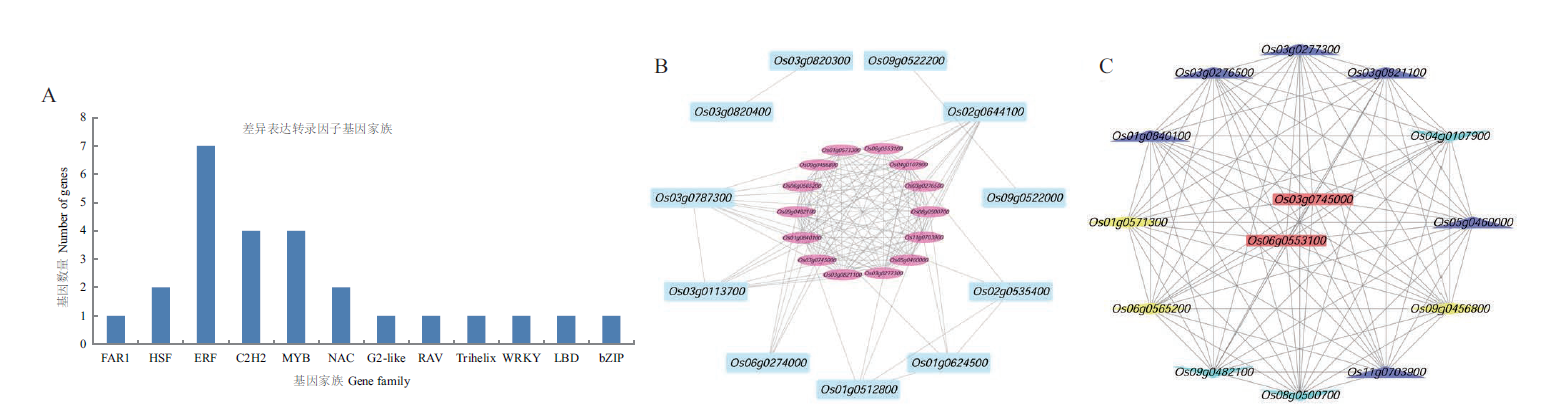
图3 相同差异表达基因分类、重要模块筛选和关键TF筛选图 A:差异表达转录因子家族分类图;B:重要模块筛选图;C:关键转录因子筛选图:蓝色上三角形为HSP70基因;黄色菱形为HSP90基因;绿色下三角为TF基因;红色长方形为关键Hsf基因
Fig. 3 Classification of the identical differentially expressed genes,important module and key TF screening A:Classification of differentially expressed TF families. B:Screening diagram of important modules. C:Screening map of key TFs:HSP70 gene is shown in the blue triangle. The yellow diamond is HSP90 gene. The green lower triangle is TF gene. The red rectangle is the key Hsf gene
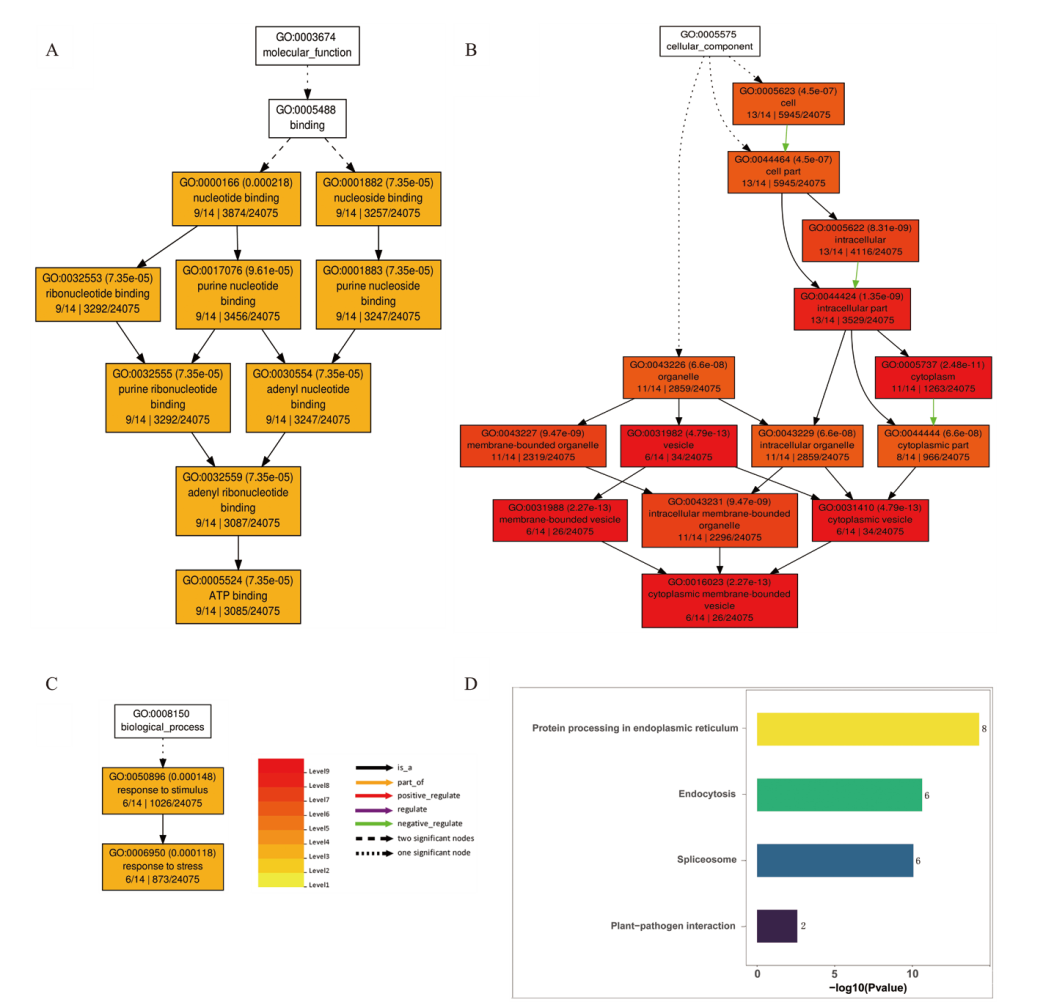
图4 重要模块基因GO和KEGG富集图 A:生物功能GO富集分析;B:细胞组分GO富集分析;C:生物过程GO富集分析;D:KEGG富集分析
Fig. 4 GO and KEGG enrichment map of important module genes A:GO enrichment analysis of biological function. B:GO enrichment analysis of cell components. C:GO enrichment analysis of biological process. D:KEGG enrichment analysis

图5 盐胁迫相关基因表达验证 A:已报道盐胁迫相关基因;B:两个关键TF基因
Fig. 5 Expression verification of salt stress related genes A:Reported genes related to salt stress. B:Two key TF genes
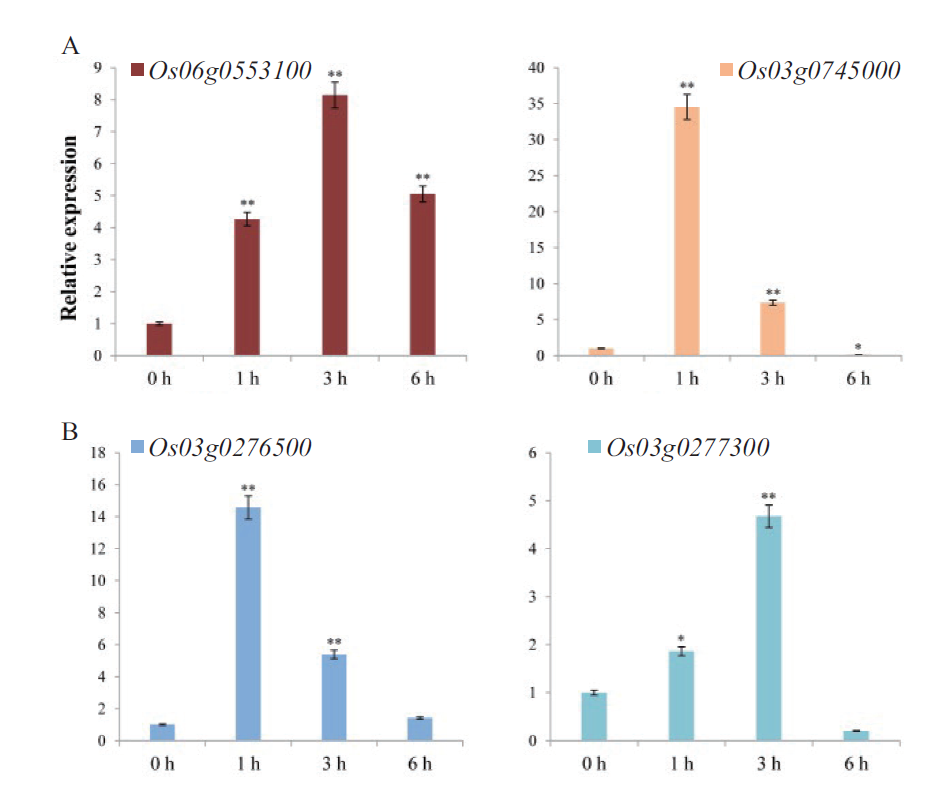
图6 盐胁迫不同时段相关基因的表达检测 A:两个关键TF基因;B:两个HSP70基因
Fig. 6 Expression detection of related genes at different time points of salt stress A:Two key TF genes. B:Two HSP70 genes
| [1] | 朱建峰, 崔振荣, 吴春红, 等. 我国盐碱地绿化研究进展与展望[J]. 世界林业研究, 2018, 31(4):70-75. |
| Zhu JF, Cui ZR, Wu CH, et al. Research advances and prospect of saline and alkali land greening in China[J]. World For Res, 2018, 31(4):70-75. | |
| [2] |
Yang Y, Guo Y. Elucidating the molecular mechanisms mediating plant salt-stress responses[J]. New Phytol, 2018, 217(2):523-539.
doi: 10.1111/nph.14920 URL |
| [3] | 李彬, 王志春, 孙志高, 等. 中国盐碱地资源与可持续利用研究[J]. 干旱地区农业研究, 2005, 23(2):154-158. |
| Li B, Wang ZC, Sun ZG, et al. Resources and sustainable resource exploitation of salinized land in China[J]. Agric Res Arid Areas, 2005, 23(2):154-158. | |
| [4] |
Dai W, Wang M, Gong X, et al. The transcription factor FcWRKY40 of Fortunella crassifolia functions positively in salt tolerance through modulation of ion homeostasis and proline biosynjournal by directly regulating SOS2 and P5CS1 homologs[J]. New Phytol, 2018, 219(3):972-989.
doi: 10.1111/nph.2018.219.issue-3 URL |
| [5] |
Liang W, Ma X, Wan P, et al. Plant salt-tolerance mechanism:a review[J]. Biochem Biophys Res Commun, 2018, 495(1):286-291.
doi: 10.1016/j.bbrc.2017.11.043 URL |
| [6] |
Duan J, Cai W. OsLEA3-2, an abiotic stress induced gene of rice plays a key role in salt and drought tolerance[J]. PLoS One, 2012, 7(9):e45117.
doi: 10.1371/journal.pone.0045117 URL |
| [7] |
Ganguly M, Datta K, Roychoudhury A, et al. Overexpression of Rab16A gene in indica rice variety for generating enhanced salt tolerance[J]. Plant Signal Behav, 2012, 7(4):502-509.
doi: 10.4161/psb.19646 URL |
| [8] | Baillo EH, Kimotho RN, Zhang ZB, et al. Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement[J]. Genes(Basel), 2019, 10(10):771. |
| [9] |
Yamamoto M, Uji S, Sugiyama T, et al. ERdj3B-mediated quality control maintains anther development at high temperatures[J]. Plant Physiol, 2020, 182(4):1979-1990.
doi: 10.1104/pp.19.01356 pmid: 31980572 |
| [10] |
Li X, Zheng H, Shi L, et al. Stress-seventy subfamily A 4, a member of HSP70, confers yeast cadmium tolerance in the loss of mitochondria pyruvate carrier 1[J]. Plant Signal Behav, 2020, 15(2):1719312.
doi: 10.1080/15592324.2020.1719312 URL |
| [11] |
Tiwari LD, Khungar L, Grover A. AtHsc70-1 negatively regulates the basal heat tolerance in Arabidopsis thaliana through affecting the activity of HsfAs and Hsp101[J]. Plant J, 2020, 103(6):2069-2083.
doi: 10.1111/tpj.v103.6 URL |
| [12] |
Biswas S, Islam MN, Sarker S, et al. Overexpression of heterotrimeric G protein beta subunit gene(OsRGB1)confers both heat and salinity stress tolerance in rice[J]. Plant Physiol Biochem, 2019, 144:334-344.
doi: 10.1016/j.plaphy.2019.10.005 URL |
| [13] |
Leng L, Liang Q, Jiang J, et al. A subclass of HSP70s regulate development and abiotic stress responses in Arabidopsis thaliana[J]. J Plant Res, 2017, 130(2):349-363.
doi: 10.1007/s10265-016-0900-6 URL |
| [14] |
Mittal D, Madhyastha DA, Grover A. Gene expression analysis in response to low and high temperature and oxidative stresses in rice:combination of stresses evokes different transcriptional changes as against stresses applied individually[J]. Plant Sci, 2012, 197:102-113.
doi: 10.1016/j.plantsci.2012.09.008 URL |
| [15] |
Chauhan H, Khurana N, Agarwal P, et al. Heat shock factors in rice(Oryza sativa L.):genome-wide expression analysis during reproductive development and abiotic stress[J]. Mol Genet Genom, 2011, 286(2):171.
doi: 10.1007/s00438-011-0638-8 URL |
| [16] |
Mittal D, Enoki Y, Lavania D, et al. Binding affinities and interactions among different heat shock element types and heat shock factors in rice(Oryza sativa L.)[J]. FEBS J, 2011, 278(17):3076-3085.
doi: 10.1111/j.1742-4658.2011.08229.x URL |
| [17] |
Singh A, Mittal D, Lavania D, et al. OsHsfA2c and OsHsfB4b are involved in the transcriptional regulation of cytoplasmic OsClpB(Hsp100)gene in rice(Oryza sativa L.)[J]. Cell Stress Chaperones, 2012, 17(2):243-254.
doi: 10.1007/s12192-011-0303-5 URL |
| [18] |
Yokotani N, Ichikawa T, Kondou Y, et al. Expression of rice heat stress transcription factor OsHsfA2e enhances tolerance to environmental stresses in transgenic Arabidopsis[J]. Planta, 2008, 227(5):957-967.
doi: 10.1007/s00425-007-0670-4 pmid: 18064488 |
| [1] | 林红妍, 郭晓蕊, 刘迪, 李慧, 陆海. 转录组分析转录因子AtbHLH68调控细胞壁发育的分子机制[J]. 生物技术通报, 2023, 39(9): 105-116. |
| [2] | 王子颖, 龙晨洁, 范兆宇, 张蕾. 利用酵母双杂交系统筛选水稻中与OsCRK5互作蛋白[J]. 生物技术通报, 2023, 39(9): 117-125. |
| [3] | 黄小龙, 孙贵连, 马丹丹, 闫慧清. 水稻幼苗酵母单杂文库构建及LAZY1上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 126-135. |
| [4] | 苗永美, 苗翠苹, 于庆才. 枯草芽孢杆菌BBs-27发酵液性质及脂肽对黄色镰刀菌的抑菌作用[J]. 生物技术通报, 2023, 39(9): 255-267. |
| [5] | 李雪琪, 张素杰, 于曼, 黄金光, 周焕斌. 基于CRISPR/CasX介导的水稻基因组编辑技术的建立[J]. 生物技术通报, 2023, 39(9): 40-48. |
| [6] | 吴元明, 林佳怡, 柳雨汐, 李丹婷, 张宗琼, 郑晓明, 逄洪波. 基于BSA-seq和RNA-seq挖掘水稻株高相关QTL[J]. 生物技术通报, 2023, 39(8): 173-184. |
| [7] | 付钰, 贾瑞瑞, 何荷, 王良桂, 杨秀莲. 两种砧木楸树嫁接苗生长差异及转录组比较分析[J]. 生物技术通报, 2023, 39(8): 251-261. |
| [8] | 姚莎莎, 王晶晶, 王俊杰, 梁卫红. 植物激素信号通路调控水稻粒型的分子机制[J]. 生物技术通报, 2023, 39(8): 80-90. |
| [9] | 王帅, 冯宇梅, 白苗, 杜维俊, 岳爱琴. 大豆GmHMGR基因响应外源激素及非生物胁迫功能研究[J]. 生物技术通报, 2023, 39(7): 131-142. |
| [10] | 魏茜雅, 秦中维, 梁腊梅, 林欣琪, 李映志. 褪黑素种子引发处理提高朝天椒耐盐性的作用机制[J]. 生物技术通报, 2023, 39(7): 160-172. |
| [11] | 李宇, 李素贞, 陈茹梅, 卢海强. 植物bHLH转录因子调控铁稳态的研究进展[J]. 生物技术通报, 2023, 39(7): 26-36. |
| [12] | 赵金玲, 安磊, 任晓亮. 单细胞转录组测序技术及其在秀丽隐杆线虫中的应用[J]. 生物技术通报, 2023, 39(6): 158-170. |
| [13] | 孔德真, 段震宇, 王刚, 张鑫, 席琳乔. 盐、碱胁迫下高丹草苗期生理特征及转录组学分析[J]. 生物技术通报, 2023, 39(6): 199-207. |
| [14] | 刘辉, 卢扬, 叶夕苗, 周帅, 李俊, 唐健波, 陈恩发. 外源硫诱导苦荞镉胁迫响应的比较转录组学分析[J]. 生物技术通报, 2023, 39(5): 177-191. |
| [15] | 任沛东, 彭健玲, 刘圣航, 姚姿婷, 朱桂宁, 陆光涛, 李瑞芳. 沙福芽孢杆菌GX-H6的分离鉴定及对水稻细菌性条斑病的防病效果[J]. 生物技术通报, 2023, 39(5): 243-253. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
