








生物技术通报 ›› 2022, Vol. 38 ›› Issue (4): 253-260.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0755
牛馨1( ), 张莹1, 王茂军1, 刘文龙2, 路福平1, 李玉1(
), 张莹1, 王茂军1, 刘文龙2, 路福平1, 李玉1( )
)
收稿日期:2021-06-10
出版日期:2022-04-26
发布日期:2022-05-06
通讯作者:
李玉,女,博士,教授,研究方向:工业酶制剂开发;E-mail: liyu@tust.edu.cn作者简介:牛馨,女,硕士研究生,研究方向:发酵工程;E-mail: niuxin0968@outlook.com
基金资助:
NIU Xin1( ), ZHANG Ying1, WANG Mao-jun1, LIU Wen-long2, LU Fu-ping1, LI Yu1(
), ZHANG Ying1, WANG Mao-jun1, LIU Wen-long2, LU Fu-ping1, LI Yu1( )
)
Received:2021-06-10
Published:2022-04-26
Online:2022-05-06
摘要:
为探究基因组上不同整合位点对外源碱性蛋白酶表达的影响,基于解淀粉芽胞杆菌TCCC 111018基因组信息,通过预测基因组上复制起始位点OriC确定了yaah基因整合位点;与此同时,通过分析6个胞外蛋白酶基因(epr、vpr、mpr、apr、bpr和nprE)单独缺失后对外源碱性蛋白酶酶活力的影响,并对发酵上清液中主要分泌蛋白的分析和质谱鉴定,确定了nprE和amyE基因位置为另外两个整合位点。在确定的3个整合位点处分别整合克劳氏芽胞杆菌来源的碱性蛋白酶基因aprE,并对整合菌株的碱性蛋白酶酶活力进行对比分析,结果表明整合菌株解淀粉芽胞杆菌18-ΔY∷aprE 和18-ΔN∷aprE碱性蛋白酶酶活力分别达到784.36 U/mL和1 289.09 U/mL,分别比原始菌株提高了15%和88.9%,实时荧光定量PCR和SDS-PAGE凝胶电泳表明18-ΔN∷aprE的碱性蛋白酶的转录水平和表达量均最高,这表明在nprE基因位点整合碱性蛋白酶基因可以有效提高碱性蛋白酶表达量,为进一步构建工业酶生产菌株奠定了基础。
牛馨, 张莹, 王茂军, 刘文龙, 路福平, 李玉. 解淀粉芽胞杆菌不同整合位点对外源碱性蛋白酶表达的影响[J]. 生物技术通报, 2022, 38(4): 253-260.
NIU Xin, ZHANG Ying, WANG Mao-jun, LIU Wen-long, LU Fu-ping, LI Yu. Effects of Different Integration Sites on the Expression of Exogenous Alkaline Protease in Bacillus amyloliquefaciens[J]. Biotechnology Bulletin, 2022, 38(4): 253-260.
| 菌株/质粒Strain/Plasmid | 特性/目的Characteristics/Purpose | 来源Source |
|---|---|---|
| 菌株Strain | ||
| E.coli EC135 pM.Bam | 对质粒DNA进行甲基化修饰 Plasmid DNA methylation modifcation | 中科院微生物研究所 Institute of Microbiology,Chinese Academy of Sciences |
| E.coli JM109 | 用于敲除质粒的构建Construction of knockout vectors | 本实验室Author’s lab |
| B.amyloliquefaciens111018 | 出发菌株Parent host | 本实验室Author’s lab |
| B.amyloliquefaciens18-ΔM | mpr基因缺失mpr gene deletion | 本研究This work |
| B.amyloliquefaciens18-ΔB | bpr基因缺失bpr gene deletion | 本研究This work |
| B.amyloliquefaciens18-ΔE | epr基因缺失epr gene deletion | 本研究This work |
| B.amyloliquefacien18-ΔV | vpr基因缺失vpr gene deletion | 本研究This work |
| B.amyloliquefaciens18-ΔN | nprE基因缺失nprE gene deletion | 本研究This work |
| B.amyloliquefaciens18-ΔA | apr基因缺失apr gene deletion | 本研究This work |
| B.amyloliquefaciens18-ΔN∷aprE | 在nprE位置整合aprE Integration of aprE in nprE site | 本研究This work |
| B.amyloliquefaciens18-Δα∷aprE | 在amyE位置整合aprE Integration of aprE in amyE site | 本研究This work |
| B.amyloliquefaciens18-ΔY∷aprE | 在yaah位置整合aprEIntegration of aprE in yaah site | 本研究This work |
| 质粒Plasmid | ||
| pWH-T2 | 穿梭表达载体Shuttle expression vector | 湖北大学Hubei University |
| pLY-2 | aprE表达载体aprE expression cassette | 本实验室Author’s lab |
表1 研究中使用的菌株和质粒
Table 1 Bacterial strains and plasmids used in this study
| 菌株/质粒Strain/Plasmid | 特性/目的Characteristics/Purpose | 来源Source |
|---|---|---|
| 菌株Strain | ||
| E.coli EC135 pM.Bam | 对质粒DNA进行甲基化修饰 Plasmid DNA methylation modifcation | 中科院微生物研究所 Institute of Microbiology,Chinese Academy of Sciences |
| E.coli JM109 | 用于敲除质粒的构建Construction of knockout vectors | 本实验室Author’s lab |
| B.amyloliquefaciens111018 | 出发菌株Parent host | 本实验室Author’s lab |
| B.amyloliquefaciens18-ΔM | mpr基因缺失mpr gene deletion | 本研究This work |
| B.amyloliquefaciens18-ΔB | bpr基因缺失bpr gene deletion | 本研究This work |
| B.amyloliquefaciens18-ΔE | epr基因缺失epr gene deletion | 本研究This work |
| B.amyloliquefacien18-ΔV | vpr基因缺失vpr gene deletion | 本研究This work |
| B.amyloliquefaciens18-ΔN | nprE基因缺失nprE gene deletion | 本研究This work |
| B.amyloliquefaciens18-ΔA | apr基因缺失apr gene deletion | 本研究This work |
| B.amyloliquefaciens18-ΔN∷aprE | 在nprE位置整合aprE Integration of aprE in nprE site | 本研究This work |
| B.amyloliquefaciens18-Δα∷aprE | 在amyE位置整合aprE Integration of aprE in amyE site | 本研究This work |
| B.amyloliquefaciens18-ΔY∷aprE | 在yaah位置整合aprEIntegration of aprE in yaah site | 本研究This work |
| 质粒Plasmid | ||
| pWH-T2 | 穿梭表达载体Shuttle expression vector | 湖北大学Hubei University |
| pLY-2 | aprE表达载体aprE expression cassette | 本实验室Author’s lab |
| 引物Primer | 序列Sequence(5'-3') |
|---|---|
| Up-F | TTAACGAATTCCTGCAGCCCGGGCAGAAAGACCA- TCCACAC |
| Up-R | TCATCCGCCAAAGCAGATAACGGATGATTC |
| Down-F | CAGCCCTCAGCTGACTGAAAAACCATTTATCATTG |
| Down-R | ATCCTTTGATCTTTTCTACGAGCTCCCGCTTATTAA- G ACGGC |
| djh-up-F | TAGCACACGTTTCATTGCAAATC |
| djh-up-R | CCGACTGCGCAAAAGACATAATC |
| djh-down-F | CCGACTGCGCAAAAGACATAATC |
| djh-down-R | TCTGAGCGCAGAAACGG |
| sjh-F | TAGCACACGTTTCATTGCAAATC |
| sjh-R | TCTGAGCGCAGAAACGG |
表2 vpr基因缺失的引物设计
Table 2 Primers for vpr gene deletion
| 引物Primer | 序列Sequence(5'-3') |
|---|---|
| Up-F | TTAACGAATTCCTGCAGCCCGGGCAGAAAGACCA- TCCACAC |
| Up-R | TCATCCGCCAAAGCAGATAACGGATGATTC |
| Down-F | CAGCCCTCAGCTGACTGAAAAACCATTTATCATTG |
| Down-R | ATCCTTTGATCTTTTCTACGAGCTCCCGCTTATTAA- G ACGGC |
| djh-up-F | TAGCACACGTTTCATTGCAAATC |
| djh-up-R | CCGACTGCGCAAAAGACATAATC |
| djh-down-F | CCGACTGCGCAAAAGACATAATC |
| djh-down-R | TCTGAGCGCAGAAACGG |
| sjh-F | TAGCACACGTTTCATTGCAAATC |
| sjh-R | TCTGAGCGCAGAAACGG |
| 引物Primer | 序列Sequence(5'-3') |
|---|---|
| 16S-F | GGGCTACACACGTGCTACAATGG |
| 16S-R | GTATTCACCGCGGCATGCTG |
| aprE-F | GAGCACATACCCAGGTTCAACG |
| aprE -R | GTTGCCGCTTCTGCATTGAC |
表3 用于实时荧光定量PCR的引物
Table 3 Primers for real-time fluorescent quantitative PCR
| 引物Primer | 序列Sequence(5'-3') |
|---|---|
| 16S-F | GGGCTACACACGTGCTACAATGG |
| 16S-R | GTATTCACCGCGGCATGCTG |
| aprE-F | GAGCACATACCCAGGTTCAACG |
| aprE -R | GTTGCCGCTTCTGCATTGAC |
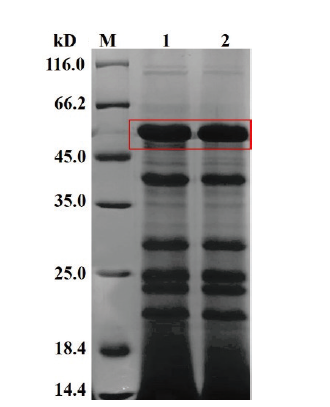
图5 TCCC 111018发酵液上清SDS-PAGE检测 M:1kb marker;1,2:TCCC 111018的发酵上清液
Fig.5 SDS-PAGE detection of TCCC 111018 fermentation broth supernatant M:1kb marker. 1 and 2:Fermentation supernatant of TCCC 111018
| RT | Mass | NCBI ID | Compound name |
|---|---|---|---|
| 1 | 58458.9 | P00692 | Alpha-amylase |
| 1.1 | 218451.1 | Q8TF72 | Protein Shroom3 |
| 1.2 | 77037.6 | O18740 | Keratin,type I cytoskeletal 9 |
| 1.3 | 56182.7 | B0TWF7 | Chromosomal replication initiator protein DnaA |
| 1.4 | 11230.8 | A5CR30 | 50S ribosomal protein L21 |
| 1.5 | 151424.3 | A4VHM3 | DNA-directed RNA polymerase subunit beta |
| 1.6 | 40412.9 | QQ4FN99 | Peptide chain release factor 1 |
| 1.7 | 18115.2 | A0NDK8 | Pro-corazonin |
| 1.8 | 100713.8 | Q65CL1 | Catenin alpha-3 |
| 1.9 | 35715.8 | G8GV69 | Dioxygenase easH |
| 2 | 22703.3 | Q6AGN3 | Nucleoside triphosphate pyrophosphatase |
| 2.1 | 33273.6 | A0R5C5 | UDP-glucose 4-epimerase |
| 2.2 | 35380.5 | Q7TVA4 | D-fructose 1,6-bisphosphatase class 2 |
| 2.3 | 107560.6 | A1A5S1 | Pre-mRNA-processing factor 6 |
| 2.4 | 75310.5 | A6TEG6 | Penicillin-binding protein activator LpoA |
| 2.5 | 229710 | P33144 | Separin |
| 2.6 | 43491.7 | A5GS98 | Ferrochelatase |
| 2.7 | 13835.7 | A9BNG3 | 30S ribosomal protein S6 |
| 2.8 | 52452.5 | A1UEN7 | Cobyric acid synthase |
| 2.9 | 48850.9 | Q28RY2 | Trigger factor |
| 3 | 40629.4 | Q5ZJ37 | Rab9 effector protein with kelch motifs |
| 3.1 | 86619.2 | P49051 | S-layer protein sap |
表4 质谱结果分析
Table 4 Analysis of mass spectrometry results
| RT | Mass | NCBI ID | Compound name |
|---|---|---|---|
| 1 | 58458.9 | P00692 | Alpha-amylase |
| 1.1 | 218451.1 | Q8TF72 | Protein Shroom3 |
| 1.2 | 77037.6 | O18740 | Keratin,type I cytoskeletal 9 |
| 1.3 | 56182.7 | B0TWF7 | Chromosomal replication initiator protein DnaA |
| 1.4 | 11230.8 | A5CR30 | 50S ribosomal protein L21 |
| 1.5 | 151424.3 | A4VHM3 | DNA-directed RNA polymerase subunit beta |
| 1.6 | 40412.9 | QQ4FN99 | Peptide chain release factor 1 |
| 1.7 | 18115.2 | A0NDK8 | Pro-corazonin |
| 1.8 | 100713.8 | Q65CL1 | Catenin alpha-3 |
| 1.9 | 35715.8 | G8GV69 | Dioxygenase easH |
| 2 | 22703.3 | Q6AGN3 | Nucleoside triphosphate pyrophosphatase |
| 2.1 | 33273.6 | A0R5C5 | UDP-glucose 4-epimerase |
| 2.2 | 35380.5 | Q7TVA4 | D-fructose 1,6-bisphosphatase class 2 |
| 2.3 | 107560.6 | A1A5S1 | Pre-mRNA-processing factor 6 |
| 2.4 | 75310.5 | A6TEG6 | Penicillin-binding protein activator LpoA |
| 2.5 | 229710 | P33144 | Separin |
| 2.6 | 43491.7 | A5GS98 | Ferrochelatase |
| 2.7 | 13835.7 | A9BNG3 | 30S ribosomal protein S6 |
| 2.8 | 52452.5 | A1UEN7 | Cobyric acid synthase |
| 2.9 | 48850.9 | Q28RY2 | Trigger factor |
| 3 | 40629.4 | Q5ZJ37 | Rab9 effector protein with kelch motifs |
| 3.1 | 86619.2 | P49051 | S-layer protein sap |
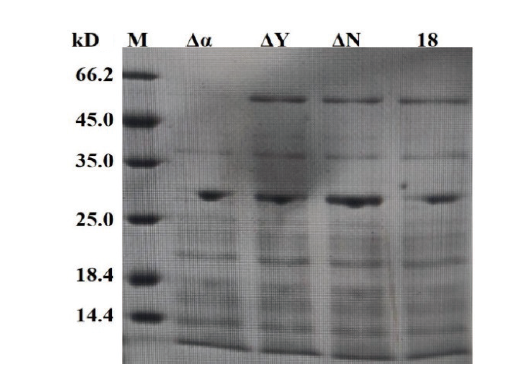
图9 整合菌株AmyE的SDS-PAGE检测 M:1 kb marker;Δα:菌株18-Δα∷aprE的发酵液上清;ΔY:菌株18-ΔY∷aprE的发酵液上清;ΔN:菌株18-ΔN∷aprE的发酵液上清;18:菌株TCCC 111018的发酵液上清
Fig.9 SDS-PAGE detection of AmyE in the integrated strains M:1 kb marker. Δα:Fermentation supernatant of strain 18-Δα∷aprE. ΔY:Fermentation supernatant of strain 18-ΔY∷aprE. ΔN:Fermentation supernatant of strain 18-ΔN∷aprE. 18:Fermentation supernatant of strain TCCC 111018
| [1] |
Ohmiya K, Tanimura S, Yashi TK, et al. Application of immobilized alkaline protease to cheese-making[J]. J Food Sci, 1979, 44(6):1584-1588.
doi: 10.1111/j.1365-2621.1979.tb09095.x URL |
| [2] |
Raval VH, Pillai S, Rawal CM, et al. Biochemical and structural characterization of a detergent-stable serine alkaline protease from seawater haloalkaliphilic bacteria[J]. Process Biochem, 2014, 49(6):955-962.
doi: 10.1016/j.procbio.2014.03.014 URL |
| [3] |
Mahmoudian M. Biocatalytic production of chiral pharmaceutical intermediates[J]. Biocatal Biotransformation, 2000, 18(2):105-118.
doi: 10.3109/10242420009015240 URL |
| [4] | Zhou C, Qin HL, Chen XJ, et al. A novel alkaline protease from alkaliphilic Idiomarina sp. C9-1 with potential application for eco-friendly enzymatic dehairing in the leather industry[J]. Sci Rep, 2018, 8(1):1-18. |
| [5] |
Omrane Benmrad M, Moujehed E, Ben Elhoul M, et al. A novel organic solvent- and detergent-stable serine alkaline protease from Trametes cingulata strain CTM10101[J]. Int J Biol Macromol, 2016, 91:961-972.
doi: 10.1016/j.ijbiomac.2016.06.025 pmid: 27296442 |
| [6] |
Baweja M, Tiwari R, Singh PK, et al. An alkaline protease from Bacillus pumilus MP 27:functional analysis of its binding model toward its applications as detergent additive[J]. Front Microbiol, 2016, 7:1195.
doi: 10.3389/fmicb.2016.01195 pmid: 27536284 |
| [7] |
Sauer C, Syvertsson S, Bohorquez LC, et al. Effect of genome position on heterologous gene expression in Bacillus subtilis:an unbiased analysis[J]. ACS Synth Biol, 2016, 5(9):942-947.
doi: 10.1021/acssynbio.6b00065 URL |
| [8] |
Bryant JA, Sellars LE, Busby SJ, et al. Chromosome position effects on gene expression in Escherichia coli K-12[J]. Nucleic Acids Res, 2014, 42(18):11383-11392.
doi: 10.1093/nar/gku828 pmid: 25209233 |
| [9] |
Kurat CF, Yeeles JTP, Patel H, et al. Chromatin controls DNA replication origin selection, lagging-strand synjournal, and replication fork rates[J]. Mol Cell, 2017, 65(1):117-130.
doi: 10.1016/j.molcel.2016.11.016 URL |
| [10] |
Couturier E, Rocha EPC. Replication-associated gene dosage effects shape the genomes of fast-growing bacteria but only for transcription and translation genes[J]. Mol Microbiol, 2006, 59(5):1506-1518.
pmid: 16468991 |
| [11] |
Zhou C, Zhou H, Li D, et al. Optimized expression and enhanced production of alkaline protease by genetically modified Bacillus licheniformis 2709[J]. Microb Cell Fact, 2020, 19(1):45.
doi: 10.1186/s12934-020-01307-2 URL |
| [12] |
Tsirigotaki A, De Geyter J, Šoštaric N, et al. Protein export through the bacterial Sec pathway[J]. Nat Rev Microbiol, 2017, 15(1):21-36.
doi: 10.1038/nrmicro.2016.161 pmid: 27890920 |
| [13] |
Mulder KCL, Bandola J, Schumann W. Construction of an artificial secYEG operon allowing high level secretion of α-amylase[J]. Protein Expr Purif, 2013, 89(1):92-96.
doi: 10.1016/j.pep.2013.02.008 URL |
| [14] |
Ren GH, Cao LC, Kong W, et al. Efficient secretion of the β-galactosidase Bgal1-3 via both tat-dependent and tat-independent pathways in Bacillus subtilis[J]. J Agric Food Chem, 2016, 64(28):5708-5716.
doi: 10.1021/acs.jafc.6b01735 URL |
| [15] |
Watzlawick H, Altenbuchner J. Multiple integration of the gene ganA into the Bacillus subtilis chromosome for enhanced β-galactosidase production using the CRISPR/Cas9 system[J]. AMB Express, 2019, 9(1):158.
doi: 10.1186/s13568-019-0884-4 pmid: 31571017 |
| [16] | Zhang G, Wang W, Deng A, et al. A mimicking-of-DNA-methylation-patterns pipeline for overcoming the restriction barrier of bacteria[J]. PLoS Genet, 2012, 8(9):e1002987. |
| [17] |
McKenney PT, Driks A, Eichenberger P. The Bacillus subtilis endospore:assembly and functions of the multilayered coat[J]. Nat Rev Microbiol, 2013, 11(1):33-44.
doi: 10.1038/nrmicro2921 pmid: 23202530 |
| [18] |
Zhang J, Xu X, Li X, et al. Reducing the cell Lysis to enhance yield of acid-stable alpha amylase by deletion of multiple peptidoglycan hydrolase-related genes in Bacillus amyloliquefaciens[J]. Int J Biol Macromol, 2021, 167:777-786.
doi: 10.1016/j.ijbiomac.2020.11.193 URL |
| [19] |
Ling Lin Fu, Zi Rong Xu, Wei Fen Li, et al. Protein secretion pathways in Bacillus subtilis:implication for optimization of heterologous protein secretion[J]. Biotechnol Adv, 2007, 25(1):1-12.
pmid: 16997527 |
| [20] |
Zhang X, Xu ZY, Liu S, et al. Improving the production of salt-tolerant glutaminase by integrating multiple copies of mglu into the protease and 16S rDNA genes of Bacillus subtilis 168[J]. Molecules, 2019, 24(3):592.
doi: 10.3390/molecules24030592 URL |
| [21] | Mo QS, Tian Y, Zhang HT, et al. Using 16S rDNA as target site for homologous recombination to improve the alkaline protease production of Bacillus alcalophilus[J]. Adv Mater Res, 2014, 886:349-354. |
| [22] |
Wang H, Zhang X, Qiu J, et al. Development of Bacillus amyloliquefaciens as a high-level recombinant protein expression system[J]. J Ind Microbiol Biotechnol, 2019, 46(1):113-123.
doi: 10.1007/s10295-018-2089-2 URL |
| [23] |
Kawamura F, Doi RH. Construction of a Bacillus subtilis double mutant deficient in extracellular alkaline and neutral proteases[J]. J Bacteriol, 1984, 160(1): 442-444.
doi: 10.1128/jb.160.1.442-444.1984 pmid: 6434524 |
| [24] |
Nakamura A, Toyama N, Kitamura A, et al. Use of a triple protease-deficient mutant of Bacillus subtilis as a host for secretion of a B. subtilis cellulase and TEM beta-lactamase[J]. Agric Biol Chem, 1991, 55(9):2367-2374.
pmid: 1368741 |
| [25] |
Abe S, Yasumura A, Tanaka T. Regulation of Bacillus subtilis aprE expression by glnA through inhibition of scoC and σ D -dependent degR expression[J]. J Bacteriol, 2009, 191(9):3050-3058.
doi: 10.1128/JB.00049-09 URL |
| [26] |
Kada S, Ishikawa A, Ohshima Y, et al. Alkaline serine protease AprE plays an essential role in poly-γ-glutamate production during natto fermentation[J]. Biosci Biotechnol Biochem, 2013, 77(4):802-809.
doi: 10.1271/bbb.120965 URL |
| [27] |
Gupta R, Gigras P, Mohapatra H, et al. Microbial α-amylases:a biotechnological perspective[J]. Process Biochem, 2003, 38(11):1599-1616.
doi: 10.1016/S0032-9592(03)00053-0 URL |
| [28] |
Wang P, Wang PL, Tian J, et al. A new strategy to express the extracellular α-amylase from Pyrococcus furiosus in Bacillus amyloliquefaciens[J]. Sci Rep, 2016, 6(1):1-10.
doi: 10.1038/s41598-016-0001-8 URL |
| [29] |
Reuβ DR, Altenbuchner J, Mäder U, et al. Large-scale reduction of the Bacillus subtilis genome:consequences for the transcriptional network, resource allocation, and metabolism[J]. Genome Res, 2017, 27(2):289-299.
doi: 10.1101/gr.215293.116 URL |
| [1] | 王帅, 冯宇梅, 白苗, 杜维俊, 岳爱琴. 大豆GmHMGR基因响应外源激素及非生物胁迫功能研究[J]. 生物技术通报, 2023, 39(7): 131-142. |
| [2] | 马玉倩, 孙东辉, 岳浩峰, 辛佳瑜, 刘宁, 曹志艳. 具有辅助降解纤维素功能的大斑刚毛座腔菌糖苷水解酶GH61的鉴定、异源表达及功能分析[J]. 生物技术通报, 2023, 39(4): 124-135. |
| [3] | 陈楠楠, 王春来, 蒋振忠, 焦鹏, 关淑艳, 马义勇. 玉米ZmDHN15基因在烟草中的遗传转化及抗冷性分析[J]. 生物技术通报, 2023, 39(4): 259-267. |
| [4] | 王玥, 高庆华, 董聪, 罗同阳, 王庆庆. 密码子优化的吡喃糖氧化酶基因在毕赤酵母中的表达[J]. 生物技术通报, 2022, 38(4): 269-277. |
| [5] | 杨瑞先, 刘萍, 王祖华, 阮宝硕, 汪智达. 牡丹根腐病原菌拮抗细菌抑菌活性物质分析[J]. 生物技术通报, 2022, 38(2): 57-66. |
| [6] | 王博雅, 姜勇, 黄艳, 曹颖, 胡尚连. 慈竹纤维素合酶BeCesA4的克隆及功能分析[J]. 生物技术通报, 2022, 38(11): 185-193. |
| [7] | 王小桃, 邹杭, 吴怡, 向省维, 吕华, 刘超兰, 林家富, 王欣荣, 褚以文, 宋涛. Paraglaciecola hydrolytica中新型β-琼胶酶Aga2的异源表达及酶学性质分析[J]. 生物技术通报, 2022, 38(11): 258-268. |
| [8] | 张彤彤, 郑登俞, 吴忠义, 张中保, 于荣. 玉米NF-Y转录因子基因ZmNF-YB13响应干旱和盐胁迫的功能分析[J]. 生物技术通报, 2022, 38(10): 115-123. |
| [9] | 廖兆民, 蔡俊, 林建国, 杜馨, 王常高. 黑曲霉葡萄糖氧化酶基因在毕赤酵母中的表达及产酶条件的优化[J]. 生物技术通报, 2021, 37(6): 97-107. |
| [10] | 袁媛, 王蕾, 石亚伟. 微生物来源碱性蛋白酶活性提高策略的研究进展[J]. 生物技术通报, 2021, 37(5): 231-236. |
| [11] | 李瑾, 彭可为, 潘求一, 朱哲远, 彭迪. 解淀粉芽胞杆菌HR-2的分离鉴定及对水稻稻瘟病菌的拮抗效果[J]. 生物技术通报, 2021, 37(3): 27-34. |
| [12] | 李昕悦, 张金方, 徐小健, 路福平, 李玉. 芽胞形成相关基因缺失对解淀粉芽胞杆菌生物量及胞外酶表达的影响[J]. 生物技术通报, 2021, 37(3): 35-43. |
| [13] | 刘珊, 叶伟, 朱牧孜, 李赛妮, 邓张双, 章卫民. 一种新型酰基转移酶GPAT的克隆、表达与酶学性质研究[J]. 生物技术通报, 2021, 37(11): 257-266. |
| [14] | 赵海燕, 宋晨斌, 刘正亚, 马兴荣, 尚会会, 李安华, 关现军, 王建设. 来源于Laceyella sp.的α-淀粉酶基因克隆、重组表达及酶学性质研究[J]. 生物技术通报, 2020, 36(8): 23-33. |
| [15] | 李卫娜, 申冬玲, 张煜星, 刘学通, 伊日布斯. Mangrovibacterium sp. SH-52耐热内切型海藻酸裂解酶基因的克隆及酶学鉴定[J]. 生物技术通报, 2020, 36(12): 82-90. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||