








生物技术通报 ›› 2021, Vol. 37 ›› Issue (11): 257-266.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1508
刘珊1,2( ), 叶伟2, 朱牧孜2, 李赛妮2, 邓张双1(
), 叶伟2, 朱牧孜2, 李赛妮2, 邓张双1( ), 章卫民2(
), 章卫民2( )
)
收稿日期:2020-12-13
出版日期:2021-11-26
发布日期:2021-12-03
作者简介:刘珊,女,硕士研究生,研究方向:微生物功能基因;E-mail: 基金资助:
LIU Shan1,2( ), YE Wei2, ZHU Mu-zi2, LI Sai-ni2, DENG Zhang-shuang1(
), YE Wei2, ZHU Mu-zi2, LI Sai-ni2, DENG Zhang-shuang1( ), ZHANG Wei-min2(
), ZHANG Wei-min2( )
)
Received:2020-12-13
Published:2021-11-26
Online:2021-12-03
摘要:
Lithocarpins为分离自海洋真菌Phomopsis lithocarpus的聚酮类新骨架化合物,具有开发成为新型抗肿瘤药物先导化合物的潜力。基于基因组测序和生物信息学分析预测了lithocarpins的生物合成基因簇,对其中的未知功能基因g7779进行扩增,并在大肠杆菌中进行表达,利用镍亲和层析柱进行纯化,通过质谱测序、生物信息学分析及酶活力检测等方法,对纯化的蛋白进行了功能分析。结果表明:基因g7779表达的蛋白是一个兼具较强的酰基转移酶活性和一定的谷丙转氨酶活性的新型双功能酶(glutamic-pyruvic transaminase-acyltransferase,GPAT),纯度达到98.6%。结构分析显示,该蛋白由326个氨基酸残基组成,其分子量为36 kD,其二级结构包括55.52% α-螺旋、29.45%无规则卷曲、9.82%延伸链和5.21% β-转角;酰基转移酶活性分析表明,GPAT对底物乙酰辅酶A的最适反应温度为35℃,当pH为7.0时酶活最高,在40℃时的热稳定性较好,在此温度下处理2 h后,残留酶活力仍大于80%,而在50℃下热稳定性较差,处理30 min后残留酶活力低于10%;其酶动力学常数Km = 761.57 μmol/L,最大反应速率Vmax = 29 370 μmol/(mg·min)。研究结果将为后续阐明lithocarpins生物合成途径提供分子生物学依据。
刘珊, 叶伟, 朱牧孜, 李赛妮, 邓张双, 章卫民. 一种新型酰基转移酶GPAT的克隆、表达与酶学性质研究[J]. 生物技术通报, 2021, 37(11): 257-266.
LIU Shan, YE Wei, ZHU Mu-zi, LI Sai-ni, DENG Zhang-shuang, ZHANG Wei-min. Cloning,Expression and Characterization of a Novel Acyltransferase GPAT[J]. Biotechnology Bulletin, 2021, 37(11): 257-266.
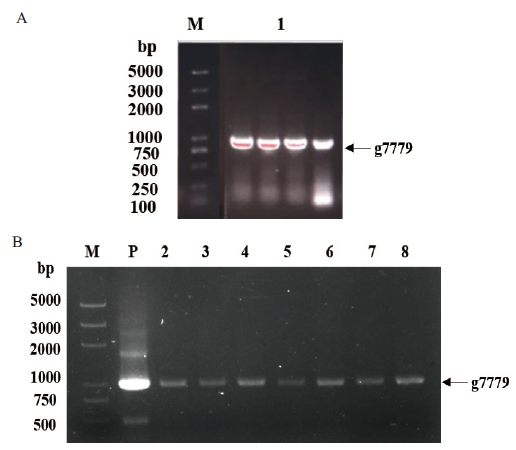
图1 基因g7779的PCR验证 A:基因g7779的克隆;B:菌液PCR验证。M:DNA marker 2K Plus;1:基因g7779;P:阳性对照;2-8:pET28a-g7779阳性单克隆
Fig.1 PCR verification of gene g7779 A:Cloning of gene g7779. B:PCR verification of bacterial solution. M:DNA marker 2K Plus. 1:Gene g7779. P:Positive control. 2-8:Positive monoclonal of pET28a-g7779

图2 蛋白GPAT的纯化与Western-Blot检测 A:蛋白GPAT表达条件的优化,M:Blue Plus Ⅱ protein marker;1、3、5、7、9、11:条件①、②、③、④、⑤、⑥的上清;2、4、6、8、10、12:条件①、②、③、④、⑤、⑥ 的沉淀。B:蛋白GPAT的纯化,M:Blue Plus Ⅱ protein marker;1:未诱导总蛋白;2:诱导后上清总蛋白;3-5:流出液;6:68 mmol/L咪唑洗脱液;7:116 mmol/L 咪唑洗脱液。C:蛋白GPAT的Western-Blot检测,M:EasySee Western marker;1:未诱导总蛋白;2:诱导后上清;3:流出液;4:68 mmol/L咪唑洗脱液;5:116 mmol/L 咪唑洗脱液
Fig.2 Purification and Western-Blot analysis of GPAT A:Optimization of the expression condition of GPAT. M:Blue Plus II protein marker. 1, 3, 5, 7, 9 and 11:Supernatant of condition ①, ②, ③, ④, ⑤ and ⑥. 2, 4, 6, 8, 10 and 12:Sediment of condition ①, ②, ③, ④, ⑤ and ⑥. B:Purification of GPAT. M:Blue Plus Ⅱ protein marker. 1:Uninduced total protein. 2:Total protein of supernatant after induction. 3-5:Binding buffer. 6:68 mmol/L imidazole eluent. 7:116 mmol/L imidazole eluent. C:Western-Blot analysis of GPAT. M:EasySee Western marker. 1:Uninduced total protein. 2:Total protein of supernatant after induction. 3:Binding buffer. 4:68 mmol/L imidazole eluent. 5:116 mmol/L imidazole eluent
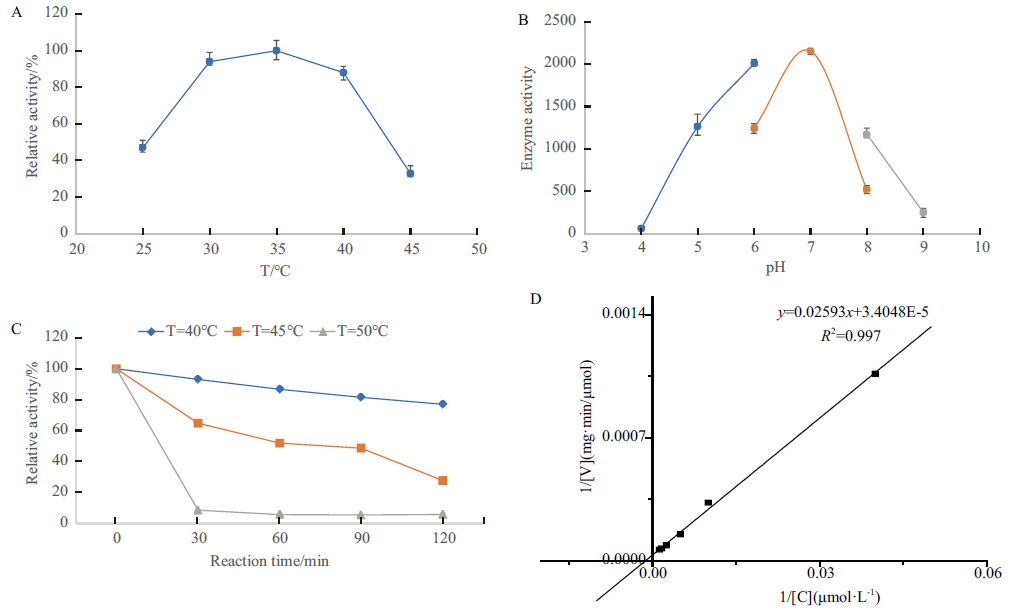
图5 GPAT的酶学性质表征 A:GPAT的最适反应温度;B:GPAT的最适反应pH;C:GPAT的热稳定性;D:GPAT的酶动力学
Fig.5 Characterization of enzymatic properties of GPAT A:Optimal reaction temperature of GPAT. B:Optimal reaction pH of GPAT. C:Thermostability of GPAT. D:Enzyme kinetics of GPAT
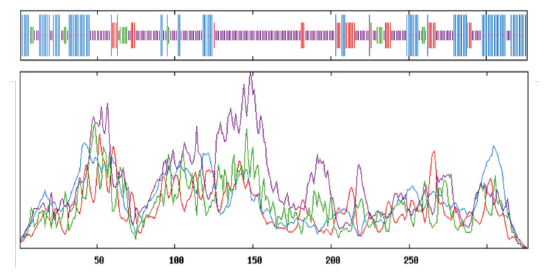
图7 蛋白GPAT的二级结构分析 竖线从长到短依次为:α-螺旋(蓝色)、延伸链(红色)、β-转角(绿色)和无规则卷曲(紫色)
Fig.7 Secondary structure analysis of GPAT The order of vertical lines from long to short are:α - helix(blue),extend strand(red),β - turn(green)and random coil(purple)
| [1] | 范东东, 孙铭娟, 王梁华, 等. 酰基转移酶研究[J]. 生命的化学, 2008, 28(6): 701-703. |
| Fan DD, Sun MJ, Wang LH, et al. Study on Acyltransferase[J]. Chemistry of Life, 2008, 28(6): 701-703. | |
| [2] |
Park YC, Shaffer CEH, Bennett GN. Microbial formation of esters[J]. Applied Microbiology and Biotechnology, 2009, 85(1): 13-25.
doi: 10.1007/s00253-009-2170-x URL |
| [3] | 张利华, 陈献忠, 陈振, 等. 热带假丝酵母肉毒碱乙酰基转移酶基因的删除及功能鉴定[J]. 食品与生物技术学报, 2018, 37(8): 880-887. |
| Zhang LH, Chen XZ, Chen Z, et al. Carnitine acetyltransferase gene disruption and function analysis in Candida Tropicalis[J]. Journal of Food Science and Biotechnology, 2018, 37(8): 880-887. | |
| [4] | 葛文雪, 陈润, 白嘉诚, 等. 结核分枝杆菌硫醇乙酰基转移酶基因敲除株的构建及其生物学特性分析[J]. 微生物与感染, 2019, 14(5): 282-288. |
| Ge WX, Chen R, Bai JC, et al. A mycothiol acetyltransferase knockout in Mycobacterium tuberculosis and its biological characteristics[J]. Journal of Microbes and Infections, 2019, 14(5): 282-288. | |
| [5] | 曹珺. 高山被孢霉中二酰甘油酰基转移酶的筛选鉴定及功能探究[D]. 江南大学, 2019. |
| Cao J. Screening and identification of diacylglycerol acyltransferase from Mortierella alpina and its effect on lipid accumulation[D]. Jiangnan University, 2019. | |
| [6] | 刘雨雨, 莫婷, 王晓晖, 等. 植物来源BAHD酰基转移酶家族研究进展[J]. 中国中药杂志, 2016, 41(12): 2175-2182. |
| Liu YY, Mo T, Wang XH, et al. Research progress of plant BAHD acyltransferase family[J]. China Journal of Chinese Materia Medica, 2016, 41(12): 2175-2182. | |
| [7] |
Xu J, Tan H, Chen Y, et al. Lithocarpins A-D:four tenellone-macrolide conjugated[4 + 2]hetero-adducts from the deep-sea derived fungus Phomopsis lithocarpus FS508[J]. Organic Chemistry Frontiers, 2018, 5: 1792-1797.
doi: 10.1039/C8QO00095F URL |
| [8] | Garvey GS, Mccormick SP, Alexander NJ, et al. Structural and functional characterization of TRI3 trichothecene 15-O-acetyltransferase from Fusarium sporotrichioides[J]. Protein Science, 2009, 18(4): 747-761. |
| [9] |
Dastjerdeh MS, Marashiyan M, Boroujeni MB, et al. In silico analysis of different signal peptides for the secretory production of recombinant human keratinocyte growth factor in Escherichia coli[J]. Computational Biology and Chemistry, 2019, 80: 225-233.
doi: S1476-9271(18)30522-X pmid: 30999249 |
| [10] | Ropón-Palacios G, Chenet-Zuta ME, Otazu K, et al. Novel multi-epitope protein containing conserved epitopes from different Leishmania species as potential vaccine candidate:integrated immunoinformatics and molecular dynamics approach[J]. Computational Biology and Chemistry, 2019, 83: 1-11. |
| [11] | Jamal, Shaheen, Sunita. Study of transmembraneous protein using bioinformatics and data mining[J]. Asian Journal of Bio Science, 2014, 1(9): 71-75. |
| [12] |
Sevindik E. Comparative and phylogenetic analysis of RuBisCO large subunit(rbcL)proteins in some Sideritis L. (Lamiaceae)species:A bioinformatic approach[J]. Genetika-Belgrade, 2019, 51(1): 69-80.
doi: 10.2298/GENSR1901069S |
| [13] | 吴姝, 伊正君, 付玉荣. 结核分枝杆菌Pst S1蛋白结构与功能的生物信息学分析[J]. 中国病原生物学杂志, 2019, 14(7): 806-810+821. |
| Wu S, Yin ZJ, Fu YR. Bioinformatic analysis of the structure and function of Pst S1 from Mycobacterium tuberculosis[J]. Journal of Pathogen Biology, 2019, 14(7): 806-810+821. | |
| [14] | 钱锦, 李丽, 常爱平, 等. 鞘氨醇单胞菌中威兰胶合成关键基因welE和welC的生物信息学分析[J]. 河南师范大学学报:自然科学版, 2020, 48(1): 82-89. |
| Qian J, Li L, Chang AP, et al. Bioinformatics analysis of the key genes welE and welC in the synjournal of Welan gum from Sphingomonas sp.[J]. Journal of Henan Normal University:Natural Science Edition, 2020, 48(1): 82-89. | |
| [15] | 方智振, 姜翠翠, 周丹蓉, 等. 基于转录组的‘三月李’及其红肉突变体ARF基因家族鉴定及分析[J]. 应用与环境生物学报, 2019, 25(6): 1388-1395. |
| Fang ZZ, Jiang CC, Zhou DR, et al. Analysis of the ARF gene family of ‘Sanyueli’ plum(Prunus salicina LindL.)and its red-fleshed mutant based on transcriptome[J]. Chinese Journal of Applied and Environmental Biology, 2019, 25(6): 1388-1395. | |
| [16] |
Qiu JJ, Wang DQ, Ma YF, et al. Identification and characterization of serine acetyltransferase encoded by the Mycobacterium tuberculosis Rv2335 gene[J]. International Journal of Molecular Medicine, 2013, 31(5): 1229-1233.
doi: 10.3892/ijmm.2013.1298 URL |
| [17] | 左泽红, 魏韬, 郭丽琼, 等. 平菇高丝氨酸乙酰基转移酶基因的克隆及异源表达优化[J]. 食品工业科技, 2019, 40(13): 64-70. |
| Zuo ZH, Wei T, Guo LQ, et al. Cloning and heterologous expression optimization of homoserine acetyltransferase gene from Pleurotus ostreatus[J]. Science and Technology of Food Industry, 2019, 40(13): 64-70. | |
| [18] | 钱玉梅, 李红侠, 李秀丽, 等. ‘赤霞珠’葡萄果实中儿茶素没食子酰基化转移酶蛋白的纯化与鉴定[J]. 果树学报, 2020, 37(8): 1132-1143. |
| Qian YM, Li HX, Li XL, et al. Purification and identification of galloyltransferase involved in catechin metabolism in ‘Cabernet Sauvignon’ grape[J]. Journal of Fruit Science, 2020, 37(8): 1132-1143. | |
| [19] | 甄军波, 刘琳琳, 杜海英, 等. 海岛棉GbGPAT2基因的克隆与表达分析[J]. 西南农业学报, 2020, 33(3): 503-508. |
| Zhen JB, Liu LL, Du HY, et al. Cloning and Expression Analysis of GbGPAT2 from Gossypium barbadense[J]. Southwest China Journal of Agricultural Sciences, 2020, 33(3): 503-508. | |
| [20] | 刘小琴, 杨岩, 吴喆瑜, 等. 多点突变提高α-L-鼠李糖苷酶热稳定性[J]. 食品与发酵工业, 2019, 45(6): 23-29. |
| Liu XQ, Yang Y, Wu ZY, et al. Improvement of thermal stability of α-L-rhamnosidase by multiple point mutation[J]. Food and Fermentation Industries, 2019, 45(6): 23-29. | |
| [21] | 刘晓彤, 邬敏辰, 殷欣, 等. 二硫键对提高木聚糖酶AoXyn11A热稳定性的作用[J]. 食品与生物技术学报, 2014, 33(10): 1038-1043. |
| Liu XT, Wu MC, Yin X, et al. Effect of disulfide bridge on thermostability improvement of the xylanase AoXyn11A[J]. Journal of Food Science and Biotechnology, 2014, 33(10): 1038-1043. | |
| [22] | 李培, 刘欣, 向彬彬, 等. 基于功能纳米材料的蛋白激酶活性分析新方法[J]. 中国科学:化学, 2015, 45(11): 1178-1193. |
|
Li P, Liu X, Xiang BB, et al. A new method for protein kinase activity analysis based on functional nanomaterials[J]. Scientia Sinica:Chimica, 2015, 45(11): 1178-1193.
doi: 10.1360/N032015-00086 URL |
| [1] | 王帅, 冯宇梅, 白苗, 杜维俊, 岳爱琴. 大豆GmHMGR基因响应外源激素及非生物胁迫功能研究[J]. 生物技术通报, 2023, 39(7): 131-142. |
| [2] | 张路阳, 韩文龙, 徐晓雯, 姚健, 李芳芳, 田效园, 张智强. 烟草TCP基因家族的鉴定及表达分析[J]. 生物技术通报, 2023, 39(6): 248-258. |
| [3] | 李敬蕊, 王育博, 解紫薇, 李畅, 吴晓蕾, 宫彬彬, 高洪波. 甜瓜PIN基因家族的鉴定及高温胁迫表达分析[J]. 生物技术通报, 2023, 39(5): 192-204. |
| [4] | 赵赛赛, 张小丹, 贾晓妍, 陶大炜, 刘可玉, 宁喜斌. 高产硝酸盐还原酶Staphylococcus simulans ZSJ6的复合诱变选育及其酶学性质研究[J]. 生物技术通报, 2023, 39(4): 103-113. |
| [5] | 马玉倩, 孙东辉, 岳浩峰, 辛佳瑜, 刘宁, 曹志艳. 具有辅助降解纤维素功能的大斑刚毛座腔菌糖苷水解酶GH61的鉴定、异源表达及功能分析[J]. 生物技术通报, 2023, 39(4): 124-135. |
| [6] | 郭三保, 宋美玲, 李灵心, 尧子钊, 桂明明, 黄胜和. 斑地锦查尔酮合酶基因及启动子的克隆与分析[J]. 生物技术通报, 2023, 39(4): 148-156. |
| [7] | 王艺清, 王涛, 韦朝领, 戴浩民, 曹士先, 孙威江, 曾雯. 茶树SMAS基因家族的鉴定及互作分析[J]. 生物技术通报, 2023, 39(4): 246-258. |
| [8] | 陈楠楠, 王春来, 蒋振忠, 焦鹏, 关淑艳, 马义勇. 玉米ZmDHN15基因在烟草中的遗传转化及抗冷性分析[J]. 生物技术通报, 2023, 39(4): 259-267. |
| [9] | 杨岚, 张晨曦, 樊学伟, 王阳光, 王春秀, 李文婷. 鸡 BMP15 基因克隆、表达模式及启动子活性分析[J]. 生物技术通报, 2023, 39(4): 304-312. |
| [10] | 杨俊钊, 张新蕊, 赵国柱, 郑菲. 新型GH5家族多结构域纤维素酶的结构与功能研究[J]. 生物技术通报, 2023, 39(4): 71-80. |
| [11] | 陈强, 邹明康, 宋家敏, 张冲, 吴隆坤. 甜瓜LBD基因家族的鉴定和果实发育进程中的表达分析[J]. 生物技术通报, 2023, 39(3): 176-183. |
| [12] | 平怀磊, 郭雪, 余潇, 宋静, 杜春, 王娟, 张怀璧. 滇牡丹PdANS的克隆、表达及与花青素含量的相关性[J]. 生物技术通报, 2023, 39(3): 206-217. |
| [13] | 苗淑楠, 高宇, 李昕儒, 蔡桂萍, 张飞, 薛金爱, 季春丽, 李润植. 大豆GmPDAT1参与油脂合成和非生物胁迫应答的功能分析[J]. 生物技术通报, 2023, 39(2): 96-106. |
| [14] | 邢媛, 宋健, 李俊怡, 郑婷婷, 刘思辰, 乔治军. 谷子AP基因家族鉴定及其对非生物胁迫的响应分析[J]. 生物技术通报, 2023, 39(11): 238-251. |
| [15] | 陈楚怡, 杨小梅, 陈胜艳, 陈斌, 岳莉然. ABA和干旱胁迫下菊花脑ZF-HD基因家族的表达分析[J]. 生物技术通报, 2023, 39(11): 270-282. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||