








生物技术通报 ›› 2022, Vol. 38 ›› Issue (6): 272-278.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1432
刘静静( ), 刘晓蕊, 李琳, 王盈, 杨海元(
), 刘晓蕊, 李琳, 王盈, 杨海元( ), 戴一凡(
), 戴一凡( )
)
收稿日期:2021-11-15
出版日期:2022-06-26
发布日期:2022-07-11
作者简介:刘静静,女,硕士研究生,研究方向:转基因大动物;E-mail: 基金资助:
LIU Jing-jing( ), LIU Xiao-rui, LI Lin, WANG Ying, YANG Hai-yuan(
), LIU Xiao-rui, LI Lin, WANG Ying, YANG Hai-yuan( ), DAI Yi-fan(
), DAI Yi-fan( )
)
Received:2021-11-15
Published:2022-06-26
Online:2022-07-11
摘要:
利用CRISPR/Cas9系统建立催产素受体(oxytocin receptor,OXTR)基因敲除的巴马猪胎儿成纤维细胞系(porcine fetal fibroblasts,PFFs),为构建OXTR基因敲除巴马猪模型提供供体细胞。首先,对人和猪的OXTR基因进行了生物学信息分析。其次,利用在线设计工具设计了2个靶向猪OXTR外显子区的sgRNA,并克隆至pX330骨架质粒中,最后将打靶质粒和Neomycin抗性质粒共转染至PFFs中,用G418药物筛选出抗性单克隆细胞,测序鉴定其基因型。生物信息学分析显示人和猪OXTR基因进化距离较近,氨基酸序列一致性达到91%,相似性为93%,三维结构的RMSD值为0.009。成功构建OXTR基因的Cas9/sgRNA打靶载体,转染PFFs细胞后,药物筛选获得OXTR基因敲除的单克隆细胞并测序证实了基因突变型。人和猪OXTR基因具有高度同源性;构建的Cas9/sgRNA表达载体高效实现了OXTR基因编辑,成功获得基因敲除的单细胞克隆,为后续构建OXTR敲除的巴马猪模型奠定了前期基础。
刘静静, 刘晓蕊, 李琳, 王盈, 杨海元, 戴一凡. 利用CRISPR/Cas9技术建立OXTR基因敲除猪胎儿成纤维细胞系[J]. 生物技术通报, 2022, 38(6): 272-278.
LIU Jing-jing, LIU Xiao-rui, LI Lin, WANG Ying, YANG Hai-yuan, DAI Yi-fan. Establishment of Porcine Fetal Fibroblasts with OXTR-knockout Using CRISPR/Cas9[J]. Biotechnology Bulletin, 2022, 38(6): 272-278.
| 名称 Primer name | 引物序列 Primer sequence(5'-3') | 产物大小 Fragment length |
|---|---|---|
| OXTR-F | ACCTGCCCAAGAAGTCTCAG | 748 bp |
| OXTR-R | CAGCAGCAGGTAAGTGGAAG |
表1 OXTR基因exon 2序列扩增引物
Table 1 Amplification primers for exon 2 sequene of OXTR gene
| 名称 Primer name | 引物序列 Primer sequence(5'-3') | 产物大小 Fragment length |
|---|---|---|
| OXTR-F | ACCTGCCCAAGAAGTCTCAG | 748 bp |
| OXTR-R | CAGCAGCAGGTAAGTGGAAG |

图2 人/猪OXTR的一级、二级、三级结构分析 A:人/猪OXTR氨基酸序列同源性分析,相同氨基酸用蓝色阴影表示;B:人/猪蛋白质二级结构,红色表示a螺旋,绿色表示β折叠,蓝色表示β转角;C:人/猪蛋白质三维建模结果,红色为人类OXTR结构,绿色为猪OXTR结构
Fig.2 Analysis of primary,secondary,and tertiary structures of OXTR between humans and pigs A:Amino acid sequence homology analysis of human and pig OXTR,and the identical amino acids are blue shaded. B:Secondary structures of human and pig proteins. Red indicates alpha-helix;green indicates beta-sheet;and blue indicates beta-turn. C:Tertiary structures of human and pig proteins. Red indicates the structure of human OXTR,and green indicates the structure of pig OXTR
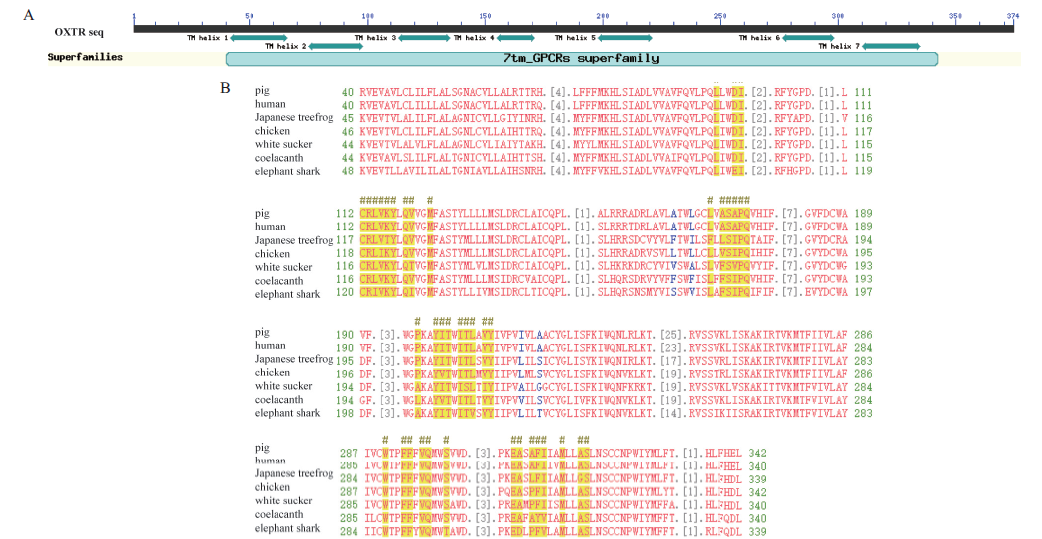
图3 猪OXTR关键残基和结构域的鉴别 A:猪OXTR的结构域和催化残基示意图;B:OXTR的多重序列比对结果,#表示与配体结合的关键氨基酸残基
Fig. 3 Identification of domains and key residues of porcine OXTR A:Schematic diagram of domains and catalytic residues of pig OXTR. B:Multiple sequence alignment of OXTR,# denotes critical amino acid residues binding to ligand
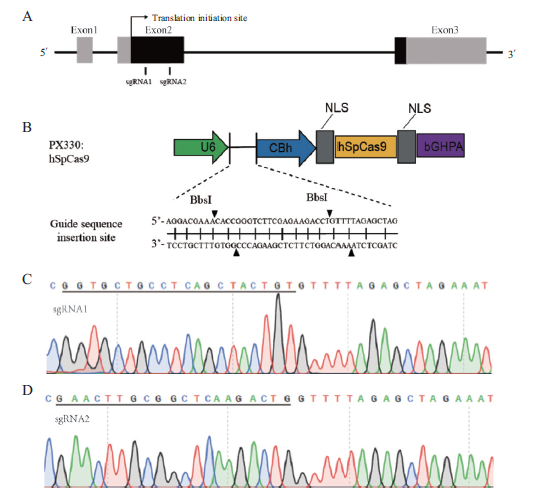
图4 OXTR基因靶点和重组载体测序 A:巴马猪OXTR基因靶点示意图,灰色表示非编码区,黑色表示编码区;B:pX330载体示意;C、D:重组载体示意图,黑色划线处为插入序列
Fig.4 Target of OXTR gene and sequencing of recombinant vectors A:Schematic diagram of OXTR gene target in Bama miniature pig. Gray indicates the non-coding area and black indicates the coding areas. B:Schematic diagram of pX330 vector. C and D:Sequencing map of recombinant vector. Black underlined is the insertion sequence
| sgRNA | 序列Sequence(5'-3') |
|---|---|
| sgRNA-1-F | CACCGGTGCTGCCTCAGCTACTGT |
| sgRNA-1-R | AAACACAGTAGCTGAGGCAGCACC |
| sgRNA-2-F | CACCGAACTTGCGGCTCAAGACTG |
| sgRNA-2-R | AAACCAGTCTTGAGCCGCAAGTTC |
表2 OXTR基因靶向位点的sgRNA寡核苷酸序列
Table 2 Oligonucleotide sequences of sgRNAs at OXTR targeting sites
| sgRNA | 序列Sequence(5'-3') |
|---|---|
| sgRNA-1-F | CACCGGTGCTGCCTCAGCTACTGT |
| sgRNA-1-R | AAACACAGTAGCTGAGGCAGCACC |
| sgRNA-2-F | CACCGAACTTGCGGCTCAAGACTG |
| sgRNA-2-R | AAACCAGTCTTGAGCCGCAAGTTC |
| 编号 ID name | 目的片段 Target fragment | 突变型 Indels |
|---|---|---|
| WT | CAGGTGCTGCCTCAGCTACTGTGGG//GGCAGAACTTGCGGCTCAAGACTGCG | WT |
| 1 | CAGGTGCTGCCTCAGCTAC-----// ----------------------CTGCGG | -411 bp |
| 2 | CAGGTGCTGCCTCAGCTAC-----// ----------------------CTGCGG | -411 bp |
| 3 | CAGGTGCTGCCTCAGCTAC-----// ----------------------CTGCGG | -411 bp |
| 4 | CAGGTGCTGCCTCAGCTACTG---// -------------------------CGG | -412 bp |
| 5 | CAGGTGCTGCCTCAGCTACTCTTGA//AGCGGAAGGTGATATCCCACACTGCGG | -289,+289 bp |
表3 OXTR 敲除PFFs的基因型
Table 3 Genotypes of OXTR-knockout PFFs
| 编号 ID name | 目的片段 Target fragment | 突变型 Indels |
|---|---|---|
| WT | CAGGTGCTGCCTCAGCTACTGTGGG//GGCAGAACTTGCGGCTCAAGACTGCG | WT |
| 1 | CAGGTGCTGCCTCAGCTAC-----// ----------------------CTGCGG | -411 bp |
| 2 | CAGGTGCTGCCTCAGCTAC-----// ----------------------CTGCGG | -411 bp |
| 3 | CAGGTGCTGCCTCAGCTAC-----// ----------------------CTGCGG | -411 bp |
| 4 | CAGGTGCTGCCTCAGCTACTG---// -------------------------CGG | -412 bp |
| 5 | CAGGTGCTGCCTCAGCTACTCTTGA//AGCGGAAGGTGATATCCCACACTGCGG | -289,+289 bp |
| [1] |
Park HR, Lee JM, Moon HE, et al. A short review on the current understanding of autism spectrum disorders[J]. Exp Neurobiol, 2016, 25(1):1-13.
doi: 10.5607/en.2016.25.1.1 pmid: 26924928 |
| [2] |
Feldman R, Monakhov M, Pratt M, et al. Oxytocin pathway genes:evolutionary ancient system impacting on human affiliation, sociality, and psychopathology[J]. Biol Psychiatry, 2016, 79(3):174-184.
doi: 10.1016/j.biopsych.2015.08.008 URL |
| [3] |
Tops S, Habel U, Radke S. Genetic and epigenetic regulatory mechanisms of the oxytocin receptor gene(OXTR)and the(clinical)implications for social behavior[J]. Horm Behav, 2019, 108:84-93.
doi: 10.1016/j.yhbeh.2018.03.002 URL |
| [4] |
Jurek B, Neumann ID. The oxytocin receptor:from intracellular signaling to behavior[J]. Physiol Rev, 2018, 98(3):1805-1908.
doi: 10.1152/physrev.00031.2017 URL |
| [5] | Vaidyanathan R, Hammock EAD. Oxytocin receptor gene loss influences expression of the oxytocin gene in C57BL/6J mice in a sex- and age-dependent manner[J]. J Neuroendocrinol, 2020, 32(2):e12821. |
| [6] |
Haram M, Bettella F, Brandt CL, et al. Contribution of oxytocin receptor polymorphisms to amygdala activation in schizophrenia spectrum disorders[J]. BJPsych Open, 2016, 2(6):353-358.
doi: 10.1192/bjpo.bp.116.003376 URL |
| [7] |
Uzefovsky F, Bethlehem RAI, Shamay-Tsoory S, et al. The oxytocin receptor gene predicts brain activity during an emotion recognition task in autism[J]. Mol Autism, 2019, 10:12.
doi: 10.1186/s13229-019-0258-4 pmid: 30918622 |
| [8] |
Caria A, Ciringione L, Falco S. Morphofunctional alterations of the hypothalamus and social behavior in autism spectrum disorders[J]. Brain Sci, 2020, 10(7):435.
doi: 10.3390/brainsci10070435 URL |
| [9] |
Pobbe RLH, Pearson BL, Defensor EB, et al. Oxytocin receptor knockout mice display deficits in the expression of autism-related behaviors[J]. Horm Behav, 2012, 61(3):436-444.
doi: 10.1016/j.yhbeh.2011.10.010 URL |
| [10] | Ribeiro D, Nunes AR, Gliksberg M, et al. Oxytocin receptor signalling modulates novelty recognition but not social preference in zebrafish[J]. J Neuroendocrinol, 2020, 32(4):e12834. |
| [11] |
Chang SWC, Brent LJN, Adams GK, et al. Neuroethology of primate social behavior[J]. Proc Natl Acad Sci USA, 2013, 110(Suppl 2):10387-10394.
doi: 10.1073/pnas.1301213110 URL |
| [12] | Ma YL, Wei JL, Zhang Q, et al. A genome scan for selection signatures in pigs[J]. PLoS One, 2015, 10(3):e0116850. |
| [13] |
Li MZ, Chen L, Tian SL, et al. Comprehensive variation discovery and recovery of missing sequence in the pig genome using multiple de novo assemblies[J]. Genome Res, 2017, 27(5):865-874.
doi: 10.1101/gr.207456.116 URL |
| [14] |
Zhang JF, Khazalwa EM, Abkallo HM, et al. The advancements, challenges, and future implications of the CRISPR/Cas9 system in swine research[J]. J Genet Genomics, 2021, 48(5):347-360.
doi: 10.1016/j.jgg.2021.03.015 URL |
| [15] |
Sauleau P, Lapouble E, Val-Laillet D, et al. The pig model in brain imaging and neurosurgery[J]. Animal, 2009, 3(8):1138-1151.
doi: 10.1017/S1751731109004649 pmid: 22444844 |
| [16] |
Gao MY, Zhu XL, Yang G, et al. CRISPR/Cas9-mediated gene editing in porcine models for medical research[J]. DNA Cell Biol, 2021, 40(12):1462-1475.
doi: 10.1089/dna.2020.6474 URL |
| [17] |
Yang W, Chen X, Li S, et al. Genetically modified large animal models for investigating neurodegenerative diseases[J]. Cell Biosci, 2021, 11(1):218.
doi: 10.1186/s13578-021-00729-8 URL |
| [18] |
Gimpl G, Fahrenholz F. The oxytocin receptor system:structure, function, and regulation[J]. Physiol Rev, 2001, 81(2):629-683.
pmid: 11274341 |
| [19] |
Tick B, Bolton P, Happé F, et al. Heritability of autism spectrum disorders:a meta-analysis of twin studies[J]. J Child Psychol Psychiatry, 2016, 57(5):585-595.
doi: 10.1111/jcpp.12499 URL |
| [20] |
Liu J, Nyholt DR, Magnussen P, et al. A genomewide screen for autism susceptibility loci[J]. Am J Hum Genet, 2001, 69(2):327-340.
doi: 10.1086/321980 pmid: 11452361 |
| [21] |
Meyer-Lindenberg A, Tost H. Neural mechanisms of social risk for psychiatric disorders[J]. Nat Neurosci, 2012, 15(5):663-668.
doi: 10.1038/nn.3083 pmid: 22504349 |
| [22] |
Cataldo I, Azhari A, Esposito G. A review of oxytocin and arginine-vasopressin receptors and their modulation of autism spectrum disorder[J]. Front Mol Neurosci, 2018, 11:27.
doi: 10.3389/fnmol.2018.00027 pmid: 29487501 |
| [23] |
Li Z, Yang HY, Wang Y, et al. Generation of tryptophan hydroxylase 2 gene knockout pigs by CRISPR/Cas9-mediated gene targeting[J]. J Biomed Res, 2017, 31(5):445-452.
doi: 10.7555/JBR.31.20170026 URL |
| [24] |
Yao J, Zeng HS, Zhang M, et al. OSBPL2-disrupted pigs recapitulate dual features of human hearing loss and hypercholesterolaemia[J]. J Genet Genomics, 2019, 46(8):379-387.
doi: 10.1016/j.jgg.2019.06.006 URL |
| [25] |
Denes CE, Cole AJ, Aksoy YA, et al. Approaches to enhance precise CRISPR/Cas9-mediated genome editing[J]. Int J Mol Sci, 2021, 22(16):8571.
doi: 10.3390/ijms22168571 URL |
| [1] | 陈小玲, 廖东庆, 黄尚飞, 陈英, 芦志龙, 陈东. 利用CRISPR/Cas9系统改造酿酒酵母的研究进展[J]. 生物技术通报, 2023, 39(8): 148-158. |
| [2] | 杨玉梅, 张坤晓. 应用CRISPR/Cas9技术建立ERK激酶相分离荧光探针定点整合的稳定细胞株[J]. 生物技术通报, 2023, 39(8): 159-164. |
| [3] | 施炜涛, 姚春鹏, 魏文康, 王蕾, 房元杰, 仝钰洁, 马晓姣, 蒋文, 张晓爱, 邵伟. 利用CRISPR/Cas9技术构建MDH2敲除细胞株及抗呕吐毒素效应研究[J]. 生物技术通报, 2023, 39(7): 307-315. |
| [4] | 刘晓燕, 祝振亮, 史广宇, 华梓宇, 杨晨, 张涌, 刘军. 乳腺生物反应器的表达优化策略[J]. 生物技术通报, 2023, 39(5): 77-91. |
| [5] | 程静雯, 曹磊, 张艳敏, 叶倩, 陈敏, 谭文松, 赵亮. CHO细胞多基因工程改造策略的建立及应用[J]. 生物技术通报, 2023, 39(2): 283-291. |
| [6] | 黄文莉, 李香香, 周炆婷, 罗莎, 姚维嘉, 马杰, 张芬, 沈钰森, 顾宏辉, 王建升, 孙勃. 利用CRISPR/Cas9技术靶向编辑青花菜BoZDS[J]. 生物技术通报, 2023, 39(2): 80-87. |
| [7] | 王兵, 赵会纳, 余婧, 陈杰, 骆梅, 雷波. 利用CRISPR/Cas9系统研究REVOLUTA参与烟草叶芽发育的调控[J]. 生物技术通报, 2023, 39(10): 197-208. |
| [8] | 李双喜, 华进联. 抗猪繁殖与呼吸障碍综合征基因编辑猪研究进展[J]. 生物技术通报, 2023, 39(10): 50-57. |
| [9] | 林蓉, 郑月萍, 徐雪珍, 李丹丹, 郑志富. 拟南芥ACOL8基因在乙烯合成与响应中的功能分析[J]. 生物技术通报, 2023, 39(1): 157-165. |
| [10] | Olalekan Amoo, 胡利民, 翟云孤, 范楚川, 周永明. 利用基因编辑技术研究BRANCHED1参与油菜分枝过程的调控[J]. 生物技术通报, 2022, 38(4): 97-105. |
| [11] | 丁亚群, 丁宁, 谢深民, 黄梦娜, 张昱, 张勤, 姜力. Vps28基因敲除小鼠模型的构建及其对泌乳和免疫性状影响的研究[J]. 生物技术通报, 2022, 38(3): 164-172. |
| [12] | 燕炯, 冯晨毅, 高学坤, 许祥, 杨佳敏, 陈朝阳. 基于CRISPR/Cas9技术构建Plin1基因敲除小鼠模型及表型分析[J]. 生物技术通报, 2022, 38(3): 173-180. |
| [13] | 钟菁, 孙玲玲, 张姝, 蒙园, 支怡飞, 涂黎晴, 徐天鹏, 濮黎萍, 陆阳清. 应用CRISPR/Cas9技术敲除Mda5基因对新城疫及传染性法氏囊病毒复制的影响[J]. 生物技术通报, 2022, 38(11): 90-96. |
| [14] | 宗梅, 韩硕, 郭宁, 段蒙蒙, 刘凡, 王桂香. 利用真空渗透和CRISPR/Cas9系统获得非转基因菜薹突变体[J]. 生物技术通报, 2022, 38(10): 159-163. |
| [15] | 王海杰, 王成稷, 郭洋, 王云, 陈艳娟, 梁敏, 王珏, 龚慧, 沈如凌. 基于CRSIPR/Cas9技术构建凝血因子8基因敲除小鼠模型及表型验证[J]. 生物技术通报, 2022, 38(10): 273-280. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||