








生物技术通报 ›› 2022, Vol. 38 ›› Issue (9): 136-146.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0112
• 细菌耐药性专题(专题主编: 刘雅红 教授 孙坚 教授) • 上一篇 下一篇
李霁虹1,2( ), 荆玉玲2, 马桂珍1(
), 荆玉玲2, 马桂珍1( ), 郭荣君2(
), 郭荣君2( ), 李世东2
), 李世东2
收稿日期:2022-01-25
出版日期:2022-09-26
发布日期:2022-10-11
作者简介:李霁虹,女,硕士研究生,研究方向:生物化工;E-mail: 基金资助:
LI Ji-hong1,2( ), JING Yu-ling2, MA Gui-zhen1(
), JING Yu-ling2, MA Gui-zhen1( ), GUO Rong-jun2(
), GUO Rong-jun2( ), LI Shi-dong2
), LI Shi-dong2
Received:2022-01-25
Published:2022-09-26
Online:2022-10-11
摘要:
无色杆菌77(Achromobacter sp. 77)是黄瓜枯萎病菌(Fusarium oxysporum f. sp. cucumerinum,Foc)菌丝际优势细菌,可随Foc菌丝迁移且状态活跃。为明确菌株77在Foc菌丝际的定殖机理,本研究在分析菌株77全基因组构成基础上,研究了其趋化特性和耐药性。Illumina Hiseq+PacBio测序分析表明,菌株77基因组全长5 868 070 bp,仅含1条染色体,GC含量为65.89%。在KEGG、COG和GO数据库分别注释到2 696、4 862和4 212个基因。菌株77基因组中含mcp、che、mot等重要趋化基因;通过CARD数据库注释,发现含有drr、emr等抗生素耐药外排泵基因、抗生素耐药基因簇以及多种抗生素的抗性基因。抗生素敏感性实验表明,菌株77对浓度低于300 µg/mL的氯霉素、卡那霉素、氨苄青霉素以及浓度低于50 µg/mL的四环素均有抗性,是一株多重耐药菌株。采用改良的游动平板法测定菌株77的趋化特性,结果表明,菌株77对1 mmol/L对羟基苯乙酸、水杨酸、α-酮戊二酸、延胡索酸、琥珀酸、苹果酸等多种根分泌物和真菌分泌物组分具有明显的趋化响应;0.5 mmol/L的上述化合物和0.5-1 mmol/L的镰刀菌酸可促进菌株77的生长。上述研究结果说明菌株77对根分泌物或真菌分泌物具有趋化和利用能力,是连作条件下无色杆菌属细菌在枯萎病菌菌丝际丰度升高的重要原因。
李霁虹, 荆玉玲, 马桂珍, 郭荣君, 李世东. 无色杆菌77的基因组构成及其趋化和耐药特性[J]. 生物技术通报, 2022, 38(9): 136-146.
LI Ji-hong, JING Yu-ling, MA Gui-zhen, GUO Rong-jun, LI Shi-dong. Genome Construction of Achromobacter 77 and Its Characteristics on Chemotaxis and Antibiotic Resistance[J]. Biotechnology Bulletin, 2022, 38(9): 136-146.
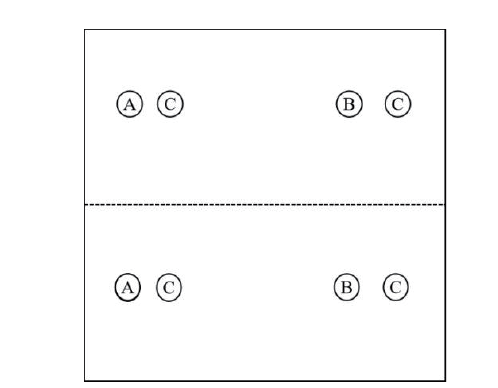
图1 细菌77对真菌分泌物及根分泌物的趋化响应示意图 上下两部分为两个平行实验。A和B分别为待测趋化化合物和无菌水的接种位置,C为细菌77菌液的接种位置
Fig. 1 Demonstration of chemotactic response of strain 77 to fungal and root exudates The plate was divided into two parts for two parallel tests of the same treatment. A and B shows the inoculation site of the candidate chemotactic compounds and sterilized water,respectively,and C shows the inoculation site of the bacterial suspension of strain 77
| 名称Name | 数据Data |
|---|---|
| 基因组大小Genome size | 5 868 070 bp |
| Scaffold条数Number of Scaffolds | 1 |
| 染色体数Number of chromosomes | 1 |
| 质粒数Number of plasmids | 0 |
| GC含量GC content | 65.89% |
| CRISPR-Cas系统数量Number of CRISPR-Cas system | 7 |
| tRNA数目Number of tRNA | 55 |
| rRNA数量Number of rRNA | 13 |
| 基因组岛数量Number of genome island | 12 |
| 基因数目Number of CDS | 5 475 |
表1 菌株77基因组组装
Table 1 Genome assembly of strain 77
| 名称Name | 数据Data |
|---|---|
| 基因组大小Genome size | 5 868 070 bp |
| Scaffold条数Number of Scaffolds | 1 |
| 染色体数Number of chromosomes | 1 |
| 质粒数Number of plasmids | 0 |
| GC含量GC content | 65.89% |
| CRISPR-Cas系统数量Number of CRISPR-Cas system | 7 |
| tRNA数目Number of tRNA | 55 |
| rRNA数量Number of rRNA | 13 |
| 基因组岛数量Number of genome island | 12 |
| 基因数目Number of CDS | 5 475 |
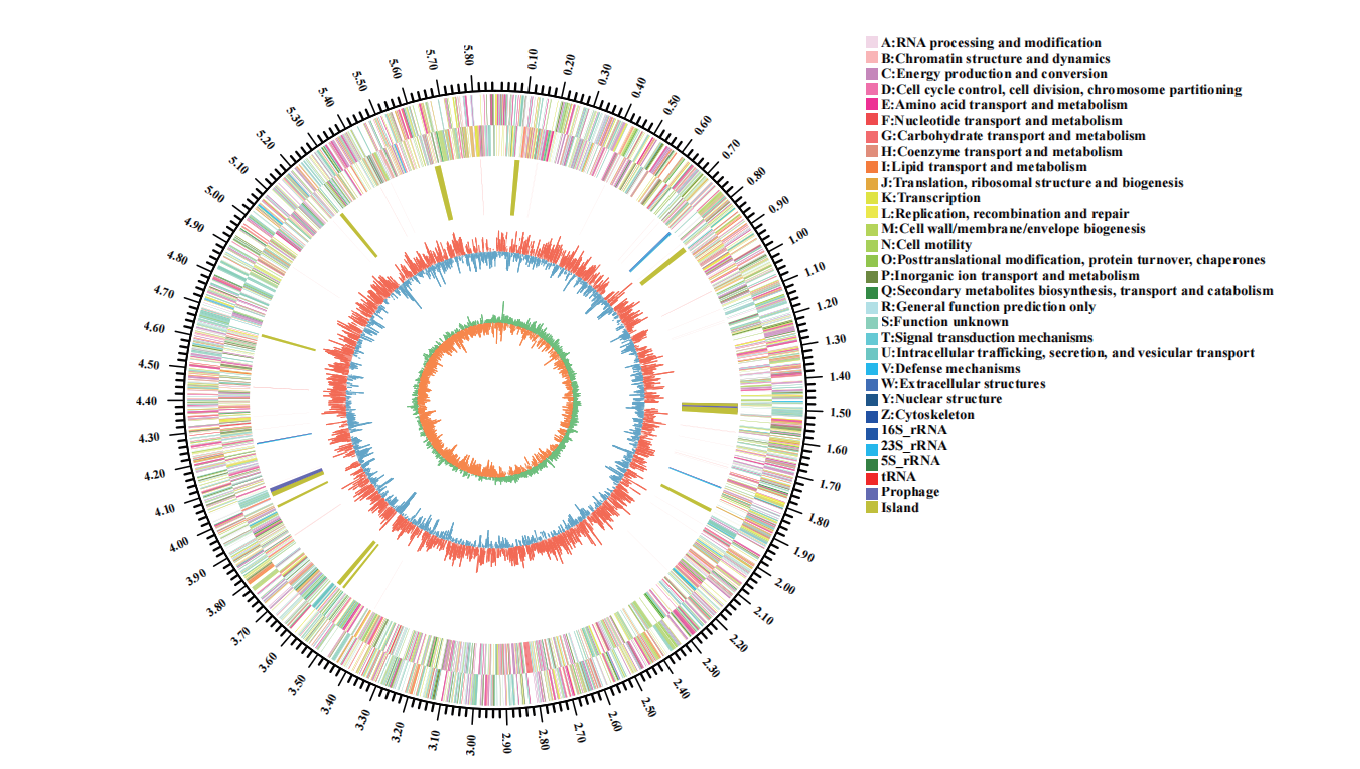
图2 菌株77的基因组圈图 最外面一圈为基因组大小的标识;第二圈和第三圈为正链、负链上的CDS,不同的颜色表示CDS不同的COG的功能分类;第四圈为rRNA和tRNA;第五圈为GC含量,向外的红色部分表示该区域GC含量高于全基因组平均GC含量,峰值越高表示与平均GC含量差值越大,向内的蓝色部分表示该区域GC含量低于全基因组平均GC含量,峰值越高表示与平均GC含量差值越大;最内一圈为GC-Skew值
Fig. 2 Circle map of the genome of strain 77 The outside circle of the map shows the genome size. The second and third circles are CDS on the positive and negative chains. Different colors indicate the functional classification of different COG of CDS. The fourth circle is rRNA and tRNA. The fifth circle is GC content. The outward red part indicates that GC content in this region is higher than the average GC content in the whole genome. The higher the peak value,the greater the difference between GC content in this region and the average GC content. The interior blue part indicates that GC content in this region is lower than the average GC content in the whole genome. The higher the peak value,the greater the difference between GC content in this region and the average GC content. The interior circle is GC-SKEW
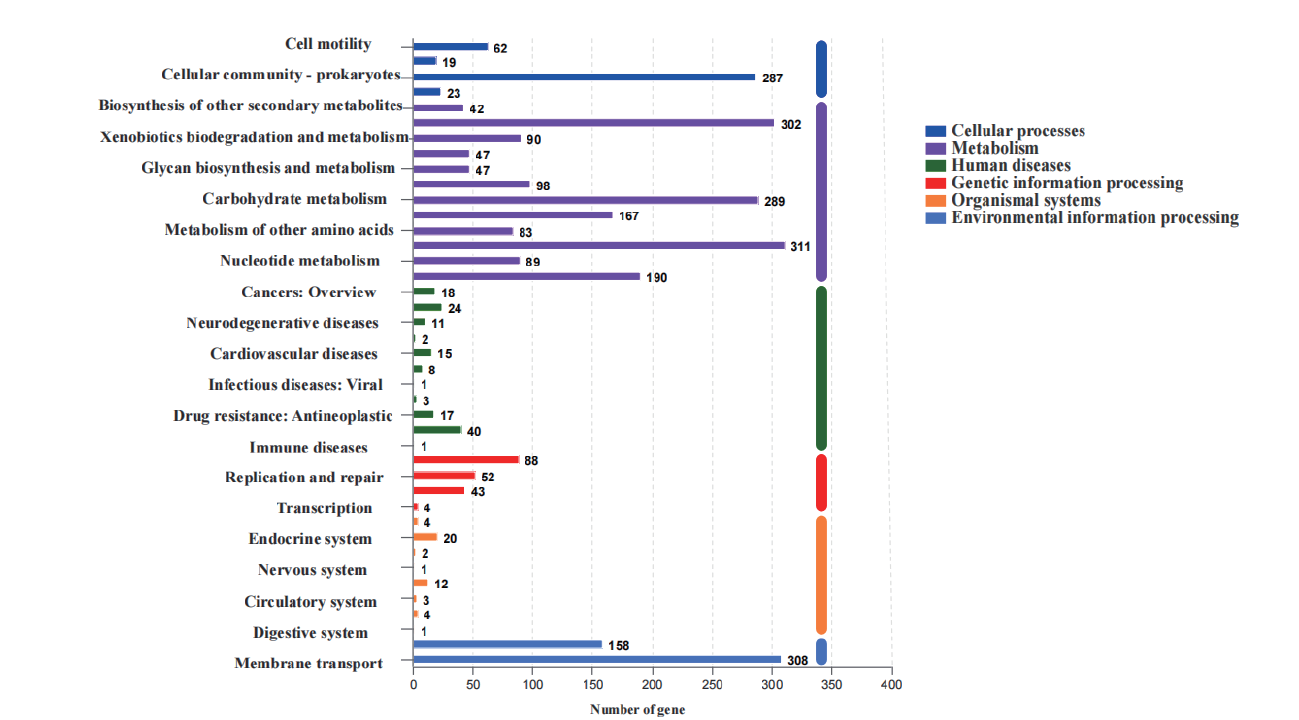
图3 菌株77基因组的KEGG注释途径 纵坐标表示KEGG pathway的level2层级分类,横坐标表示在注释该分类下基因的数目。不同柱子颜色代表KEGG pathway的level1层级分类。最右侧柱子表示不同level1分类下的基因数目。由于同一个基因可能会被注释到多个level2分类中,因此计算level1分类的基因数目时会进行去冗余处理
Fig. 3 KEGG annotated pathways of strain 77's genome The vertical coordinate represent the level2 classification of KEGG pathway,and the horizontal coordinate represents the number of genes under this classification. Different colors represent the level1 classification of KEGG pathway. The right column represents the number of genes under different level1 classifications. Since the same gene may be annotated into more than one level2 categories,de-redundancy is performed when counting the number of genes in level1 categories
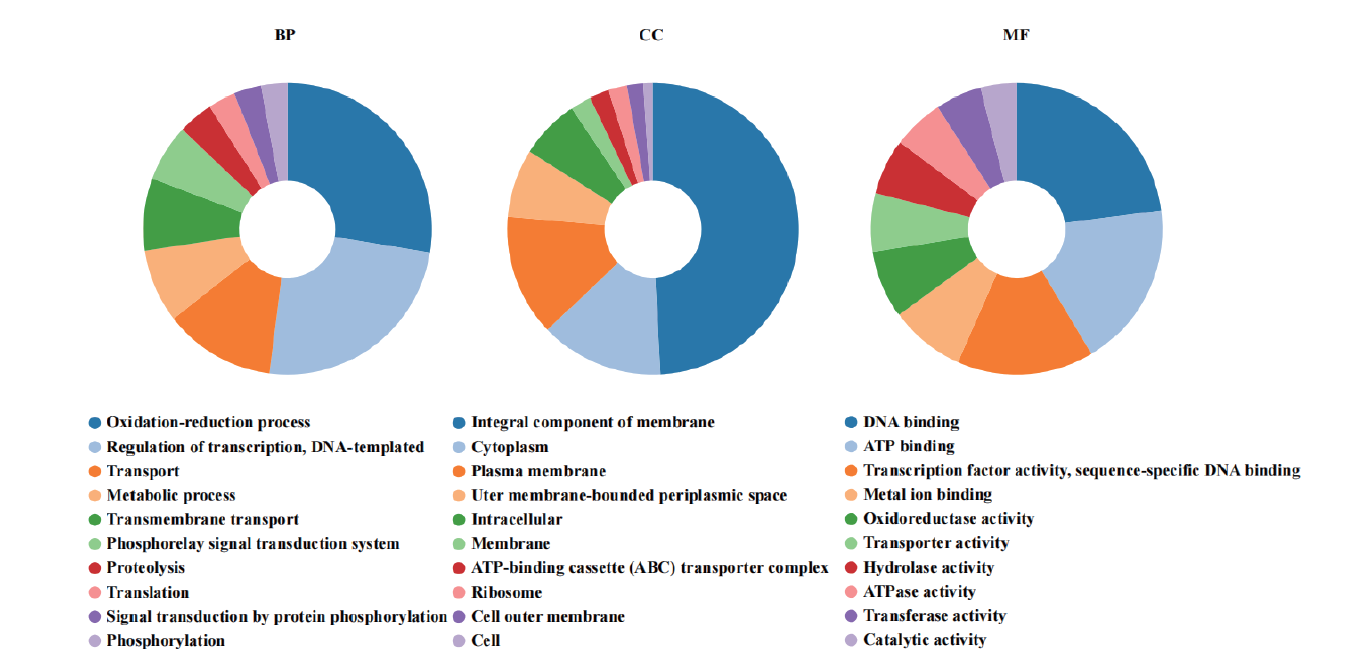
图5 菌株77基因组的GO功能注释分类 BP、CC、MF分别为GO功能中的生物过程(biological process)、细胞组分(cellular component)、分子功能(molecular function),每个饼图展示TOP 10的GO功能
Fig. 5 Classification of GO functions annotation of strain 77's genome Each category shows TOP 10 GO annotations
| GO注释GO annotation | 基因数Number of genes |
|---|---|
| 代谢过程Metabolic process | 158 |
| 三羧酸循环Tricarboxylic acid cycle | 18 |
| 苹果酸代谢Malate metabolic process | 4 |
| 细胞氨基酸分解代谢 Cellular amino acid catabolic process | 5 |
| 胁迫应答Stress response | 22 |
| 抗生素分解Antibiotic decomposition | 2 |
| 药物跨膜运输Transmembrane transport of drugs | 2 |
| 细胞氧化解毒Cellular oxidant detoxification | 33 |
| 趋化性Chemotaxis | 33 |
| 细菌型鞭毛依赖运动 Bacterial-type flagellum-dependent cell motility | 18 |
表2 菌株77基因组的GO注释中分解代谢与趋化相关基因统计
Table 2 Statistics of catabolism and chemotaxis related genes in GO annotation of strain 77's genome
| GO注释GO annotation | 基因数Number of genes |
|---|---|
| 代谢过程Metabolic process | 158 |
| 三羧酸循环Tricarboxylic acid cycle | 18 |
| 苹果酸代谢Malate metabolic process | 4 |
| 细胞氨基酸分解代谢 Cellular amino acid catabolic process | 5 |
| 胁迫应答Stress response | 22 |
| 抗生素分解Antibiotic decomposition | 2 |
| 药物跨膜运输Transmembrane transport of drugs | 2 |
| 细胞氧化解毒Cellular oxidant detoxification | 33 |
| 趋化性Chemotaxis | 33 |
| 细菌型鞭毛依赖运动 Bacterial-type flagellum-dependent cell motility | 18 |
| 基因类别 Gene classification | 基因名称 Gene name | 基因数量 Number of genes | 基因编号 Gene ID | KEGG数据库基因注释 Gene annotation in KEGG database |
|---|---|---|---|---|
| mcp | mcp | 9 | gene0255、gene0975、gene1020、gene2196、gene3307、gene3446、gene4030、gene4485、gene4827 | 甲基趋化受体蛋白Mcp |
| che | cheA | 1 | gene2189 | 感受器激酶CheA |
| cheW | 1 | gene2190 | 嘌呤结合趋化蛋白CheW | |
| cheR | 2 | gene2192、gene4468 | 甲基转移酶CheR | |
| cheB | 2 | gene2193、gene3711 | 趋化响应调节蛋白CheB | |
| cheBR | 1 | gene4467 | CheB/CheR融合蛋白 | |
| cheD | 1 | gene5023 | 趋化蛋白CheD | |
| wsp | wspF | 1 | gene3040 | 趋化响应调节蛋白WspF |
| wspD、wspB | 2 | gene3042、gene3044 | 趋化相关蛋白WspD、WspB | |
| wspC | 1 | gene3043 | 甲基转移酶WspC | |
| wspA | 1 | gene3045 | 甲基趋化受体蛋白WspA | |
| mot | motA | 1 | gene2381 | 鞭毛马达蛋白MotA |
| aer | aer | 2 | gene2219、gene2311 | 趋氧性受体蛋白Aer |
| tar | tar | 1 | gene4676 | 甲基趋化受体蛋白Ⅱ(天冬氨酸感受器受体) |
| - | - | 5 | gene0304、gene2191、gene2214、gene2215、gene2217 | 未知功能趋化蛋白 |
| - | - | 1 | gene3125 | 未知功能趋化蛋白 |
| - | - | 1 | gene3000 | 未知功能趋化蛋白 |
表3 菌株77趋化基因预测分类
Table 3 Predictive classification of chemotactic genes of strain 77
| 基因类别 Gene classification | 基因名称 Gene name | 基因数量 Number of genes | 基因编号 Gene ID | KEGG数据库基因注释 Gene annotation in KEGG database |
|---|---|---|---|---|
| mcp | mcp | 9 | gene0255、gene0975、gene1020、gene2196、gene3307、gene3446、gene4030、gene4485、gene4827 | 甲基趋化受体蛋白Mcp |
| che | cheA | 1 | gene2189 | 感受器激酶CheA |
| cheW | 1 | gene2190 | 嘌呤结合趋化蛋白CheW | |
| cheR | 2 | gene2192、gene4468 | 甲基转移酶CheR | |
| cheB | 2 | gene2193、gene3711 | 趋化响应调节蛋白CheB | |
| cheBR | 1 | gene4467 | CheB/CheR融合蛋白 | |
| cheD | 1 | gene5023 | 趋化蛋白CheD | |
| wsp | wspF | 1 | gene3040 | 趋化响应调节蛋白WspF |
| wspD、wspB | 2 | gene3042、gene3044 | 趋化相关蛋白WspD、WspB | |
| wspC | 1 | gene3043 | 甲基转移酶WspC | |
| wspA | 1 | gene3045 | 甲基趋化受体蛋白WspA | |
| mot | motA | 1 | gene2381 | 鞭毛马达蛋白MotA |
| aer | aer | 2 | gene2219、gene2311 | 趋氧性受体蛋白Aer |
| tar | tar | 1 | gene4676 | 甲基趋化受体蛋白Ⅱ(天冬氨酸感受器受体) |
| - | - | 5 | gene0304、gene2191、gene2214、gene2215、gene2217 | 未知功能趋化蛋白 |
| - | - | 1 | gene3125 | 未知功能趋化蛋白 |
| - | - | 1 | gene3000 | 未知功能趋化蛋白 |
| 分泌系统Secretion system | 数量Amount |
|---|---|
| Ⅰ型 Type Ⅰ | 1 |
| Ⅱ型 Type Ⅱ | 11 |
| Ⅲ型 Type Ⅲ | 0 |
| Ⅳ型 Type Ⅳ | 2 |
| Ⅴ型 Type Ⅴ | 0 |
| Ⅵ型 Type Ⅵ | 17 |
表4 菌株77分泌系统分类
Table 4 Classification in secretion system of strain 77
| 分泌系统Secretion system | 数量Amount |
|---|---|
| Ⅰ型 Type Ⅰ | 1 |
| Ⅱ型 Type Ⅱ | 11 |
| Ⅲ型 Type Ⅲ | 0 |
| Ⅳ型 Type Ⅳ | 2 |
| Ⅴ型 Type Ⅴ | 0 |
| Ⅵ型 Type Ⅵ | 17 |

图8 菌株77对真菌分泌物及根分泌物成分的趋化响应 红色圆点表示化合物或无菌水的点接位置,红色箭头指向菌株77因趋化响应而形成的弧形趋化圈
Fig.8 Chemotactic responses of stain 77 to fungal and root exudates The red dots indicate the inoculation site of the candidate chemotactic compounds or sterile water,and the red arrows point to the arc-shaped chemotactic circle formed by the chemotactic response of strain 77
| 化合物浓度 Concentration/ (mmol.L-1) | 真菌或根分泌物 Fungal or root exudates | 根分泌物 Root exudate | 尖孢镰刀菌分泌物 Exudate of Fusarium oxysporum | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 延胡索酸 Fumaric acid | α-酮戊二酸 α -ketoglutaric acid | 苹果酸 Malic acid | 琥珀酸 Succinic acid | 水杨酸 Salicylic acid | 对羟基苯乙酸4-Hyd-roxyphenylacetic acid | 镰刀菌酸 Fusaric acid | |||
| 0 | 0.278±0.002 d | 0.334±0.002 bc | 0.274±0.004 b | 0.285±0.012 c | 0.295±0.003 d | 0.642±0.008 b | 0.293±0.004 c | ||
| 0.5 | 0.323±0.003 b | 0.344±0.002 ab | 0.319±0.005 a | 0.315±0.004 a | 0.334±0.002 b | 0.775±0.003 a | 0.340±0.002 a | ||
| 1 | 0.351±0.001 a | 0.351±0.002 a | 0.312±0.002 a | 0.313±0.006 ab | 0.366±0.002 a | 0.760±0.019 a | 0.322±0.005 b | ||
| 2 | 0.304±0.009 c | 0.331±0.001 bc | 0.285±0.004 b | 0.298±0.011 bc | 0.317±0.006 bc | 0.049±0.002 c | 0.272±0.018 d | ||
| 4 | 0.299±0.012 c | 0.326±0.004 c | 0.272±0.006 bc | 0.291±0.002 c | 0.307±0.003 cd | 0.049±0.003 c | 0.111±0.007 e | ||
| 6 | 0.288±0.002 d | 0.308±0.002 d | 0.088±0.011 d | 0.243±0.012 d | 0.223±0.026 e | 0.053±0.002 c | 0.055±0.004 f | ||
| 8 | 0.063±0.002 e | 0.095±0.018 e | 0.052±0.002 e | 0.094±0.005 e | 0.054±0.003 f | 0.052±0.003 c | 0.054±0.005 f | ||
| 10 | 0.054±0.004 e | 0.056±0.007 f | 0.054±0.003 e | 0.058±0.002 f | 0.051±0.001 f | 0.053±0.001 c | 0.053±0.003 f | ||
表5 真菌及根分泌物组分对菌株77 OD600的影响
Table 5 Effects of fungal and root exudate components on OD600 value of strain 77
| 化合物浓度 Concentration/ (mmol.L-1) | 真菌或根分泌物 Fungal or root exudates | 根分泌物 Root exudate | 尖孢镰刀菌分泌物 Exudate of Fusarium oxysporum | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 延胡索酸 Fumaric acid | α-酮戊二酸 α -ketoglutaric acid | 苹果酸 Malic acid | 琥珀酸 Succinic acid | 水杨酸 Salicylic acid | 对羟基苯乙酸4-Hyd-roxyphenylacetic acid | 镰刀菌酸 Fusaric acid | |||
| 0 | 0.278±0.002 d | 0.334±0.002 bc | 0.274±0.004 b | 0.285±0.012 c | 0.295±0.003 d | 0.642±0.008 b | 0.293±0.004 c | ||
| 0.5 | 0.323±0.003 b | 0.344±0.002 ab | 0.319±0.005 a | 0.315±0.004 a | 0.334±0.002 b | 0.775±0.003 a | 0.340±0.002 a | ||
| 1 | 0.351±0.001 a | 0.351±0.002 a | 0.312±0.002 a | 0.313±0.006 ab | 0.366±0.002 a | 0.760±0.019 a | 0.322±0.005 b | ||
| 2 | 0.304±0.009 c | 0.331±0.001 bc | 0.285±0.004 b | 0.298±0.011 bc | 0.317±0.006 bc | 0.049±0.002 c | 0.272±0.018 d | ||
| 4 | 0.299±0.012 c | 0.326±0.004 c | 0.272±0.006 bc | 0.291±0.002 c | 0.307±0.003 cd | 0.049±0.003 c | 0.111±0.007 e | ||
| 6 | 0.288±0.002 d | 0.308±0.002 d | 0.088±0.011 d | 0.243±0.012 d | 0.223±0.026 e | 0.053±0.002 c | 0.055±0.004 f | ||
| 8 | 0.063±0.002 e | 0.095±0.018 e | 0.052±0.002 e | 0.094±0.005 e | 0.054±0.003 f | 0.052±0.003 c | 0.054±0.005 f | ||
| 10 | 0.054±0.004 e | 0.056±0.007 f | 0.054±0.003 e | 0.058±0.002 f | 0.051±0.001 f | 0.053±0.001 c | 0.053±0.003 f | ||
| [1] | Staněk M. Microorganisms in the hyphosphere of fungi. I. introduction[J]. Česká Mykologie, 1984, 38(1):1-10. |
| [2] | Deveau A, Labbé J. Mycorrhiza helper bacteria[M]// Molecular Mycorrhizal Symbiosis. Hoboken, NJ, USA: John Wiley & Sons, Inc., 2016:437-450. |
| [3] |
Boer WD, Folman LB, Summerbell RC, et al. Living in a fungal world:impact of fungi on soil bacterial niche development[J]. FEMS Microbiol Rev, 2005, 29(4):795-811.
doi: 10.1016/j.femsre.2004.11.005 URL |
| [4] |
Sun RL, Jing YL, de Boer W, et al. Dominant hyphae-associated bacteria of Fusarium oxysporum f. sp. cucumerinum in different cropping systems and insight into their functions[J]. Appl Soil Ecol, 2021, 165:103977.
doi: 10.1016/j.apsoil.2021.103977 URL |
| [5] |
Bharadwaj DP, Alström S, Lundquist PO. Interactions among Glomus irregulare, arbuscular mycorrhizal spore-associated bacteria, and plant pathogens under in vitro conditions[J]. Mycorrhiza, 2012, 22(6):437-447.
doi: 10.1007/s00572-011-0418-7 pmid: 22081167 |
| [6] |
Toljander JF, Lindahl BD, Paul LR, et al. Influence of arbuscular mycorrhizal mycelial exudates on soil bacterial growth and community structure[J]. FEMS Microbiol Ecol, 2007, 61(2):295-304.
pmid: 17535297 |
| [7] |
de Weert S, Vermeiren H, Mulders IHM, et al. Flagella-driven chemotaxis towards exudate components is an important trait for tomato root colonization by Pseudomonas fluorescens[J]. Mol Plant Microbe Interact, 2002, 15(11):1173-1180.
doi: 10.1094/MPMI.2002.15.11.1173 URL |
| [8] |
Haq IU, Zwahlen RD, Yang P, et al. The response of Paraburkholderia terrae strains to two soil fungi and the potential role of oxalate[J]. Front Microbiol, 2018, 9:989.
doi: 10.3389/fmicb.2018.00989 URL |
| [9] | 孙宁康, 江飞焰, 张林, 等. 丛枝菌根真菌Rhizophagus irregularis菌丝分泌物可诱导解磷细菌Rahnella aquatilis向菌丝移动[J]. 科学通报, 2021, 66(32):4157-4168. |
|
Sun NK, Jiang FY, Zhang L, et al. Hyphal exudates of an arbuscular mycorrhizal fungus Rhizophagus irregularis induce phosphate-solubilizing bacterium Rahnella aquatilis to swim towards its hyphae[J]. Chin Sci Bull, 2021, 66(32):4157-4168.
doi: 10.1360/TB-2021-0579 URL |
|
| [10] |
Nazir R, et al. Inhibition of mushroom formation and induction of glycerol release-ecological strategies of Burkholderia terrae BS001 to create a hospitable niche at the fungus Lyophyllum sp. strain Karsten[J]. Microb Ecol, 2013, 65(1):245-254.
doi: 10.1007/s00248-012-0100-4 URL |
| [11] |
Delcher AL, Bratke KA, Powers EC, et al. Identifying bacterial genes and endosymbiont DNA with Glimmer[J]. Bioinformatics, 2007, 23(6):673-679.
pmid: 17237039 |
| [12] | Besemer J, Borodovsky M. GeneMark:web software for gene finding in prokaryotes, eukaryotes and viruses[J]. Nucleic Acids Res, 2005, 33(Web Server issue):W451-W454. |
| [13] | Chan PP, Lowe TM. tRNAscan-SE:searching for tRNA genes in genomic sequences[J]. Methods Mol Biol, 2019, 1962:1-14. |
| [14] |
Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND[J]. Nat Methods, 2015, 12(1):59-60.
doi: 10.1038/nmeth.3176 pmid: 25402007 |
| [15] |
Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000[J]. Nucleic Acids Res, 2000, 28(1):45-48.
pmid: 10592178 |
| [16] | Jensen LJ, Julien P, Kuhn M, et al. eggNOG:automated construction and annotation of orthologous groups of genes[J]. Nucleic Acids Res, 2007, 36(suppl_1):D250-D254. |
| [17] |
Kanehisa M, Goto S. KEGG:Kyoto encyclopedia of genes and genomes[J]. Nucleic Acids Res, 2000, 28(1):27-30.
doi: 10.1093/nar/28.1.27 pmid: 10592173 |
| [18] |
Konishi H, Hio M, Kobayashi M, et al. Bacterial chemotaxis towards polysaccharide pectin by pectin-binding protein[J]. Sci Rep, 2020, 10(1):3977.
doi: 10.1038/s41598-020-60274-1 pmid: 32132546 |
| [19] |
Sampedro I, Parales RE, Krell T, et al. Pseudomonas chemotaxis[J]. FEMS Microbiol Rev, 2015, 39(1):17-46.
doi: 10.1111/j.1574-6968.1986.tb01837.x URL |
| [20] |
Feng HC, Zhang N, et al. Identification of chemotaxis compounds in root exudates and their sensing chemoreceptors in plant-growth-promoting rhizobacteria Bacillus amyloliquefaciens SQR9[J]. Mol Plant Microbe Interact, 2018, 31(10):995-1005.
doi: 10.1094/MPMI-01-18-0003-R URL |
| [21] |
Warmink JA, Nazir R, van Elsas JD. Universal and species-specific bacterial ‘fungiphiles' in the mycospheres of different basidiomycetous fungi[J]. Environ Microbiol, 2009, 11(2):300-312.
doi: 10.1111/j.1462-2920.2008.01767.x pmid: 19196267 |
| [22] |
Hou S, Larsen RW, Boudko D, et al. Myoglobin-like aerotaxis transducers in Archaea and bacteria[J]. Nature, 2000, 403(6769):540-544.
doi: 10.1038/35000570 URL |
| [23] |
Sun YP, Unestam T, et al. Exudation-reabsorption in a mycorrhizal fungus, the dynamic interface for interaction with soil and soil microorganisms[J]. Mycorrhiza, 1999, 9(3):137-144.
doi: 10.1007/s005720050298 URL |
| [24] |
de Weert S, Kuiper I, et al. Role of chemotaxis toward fusaric acid in colonization of hyphae of Fusarium oxysporum f. sp. radicis-lycopersici by Pseudomonas fluorescens WCS365[J]. Mol Plant Microbe Interact, 2004, 17(11):1185-1191.
doi: 10.1094/MPMI.2004.17.11.1185 URL |
| [25] |
Liu YP, Chen L, Wu GW, et al. Identification of root-secreted com-pounds involved in the communication between cucumber, the bene-ficial Bacillus amyloliquefaciens, and the soil-borne pathogen Fu-sarium oxysporum[J]. Mol Plant Microbe Interact, 2017, 30(1):53-62.
doi: 10.1094/MPMI-07-16-0131-R URL |
| [26] |
Emmett BD, Lévesque-Tremblay V, Harrison MJ. Conserved and reproducible bacterial communities associate with extraradical hyphae of arbuscular mycorrhizal fungi[J]. ISME J, 2021, 15(8):2276-2288.
doi: 10.1038/s41396-021-00920-2 pmid: 33649552 |
| [27] | Vila T, Nazir R, Rozental S, et al. The role of hydrophobicity and surface receptors at hyphae of Lyophyllum sp. strain Karsten in the interaction with Burkholderia terrae BS001 - implications for interactions in soil[J]. Front Microbiol, 2016, 7:1689. |
| [28] |
Haq IU, Calixto RO, Yang P, et al. Chemotaxis and adherence to fungal surfaces are key components of the behavioral response of Burkholderia terrae BS001 to two selected soil fungi[J]. FEMS Microbiol Ecol, 2016, 92(11):fiw164.
doi: 10.1093/femsec/fiw164 URL |
| [29] | 廖晓敬, 杨璐溪, 等. 几株新鞘氨醇杆菌的趋化性及其相关基因组成特点[J]. 微生物学报, 2017, 57(3):399-410. |
| Liao XJ, Yang LX, et al. Chemotaxis and characteristics of chemotactic genes in Novosphingobium strains[J]. Acta Microbiol Sin, 2017, 57(3):399-410. | |
| [30] |
Singh T, Arora DK. Motility and chemotactic response of Pseudomonas fluorescens toward chemoattractants present in the exudate of Macrophomina phaseolina[J]. Microbiol Res, 2001, 156(4):343-351.
pmid: 11770852 |
| [31] |
Palmieri D, Vitale S, Lima G, et al. A bacterial endophyte exploits chemotropism of a fungal pathogen for plant colonization[J]. Nat Commun, 2020, 11(1):5264.
doi: 10.1038/s41467-020-18994-5 pmid: 33067433 |
| [32] |
Yuan J, Zhang N, Huang QW, et al. Organic acids from root exudates of banana help root colonization of PGPR strain Bacillus amyloliquefaciens NJN-6[J]. Sci Rep, 2015, 5:13438.
doi: 10.1038/srep13438 pmid: 26299781 |
| [33] | 郭艾云, 鲍艳宇, 周启星. 土壤农药污染与细菌农药-抗生素交叉抗性研究进展[J]. 微生物学通报, 2020, 47(9):2984-2995. |
| Guo AY, Bao YY, Zhou QX. Advances in soil pesticide contamination and bacterial pesticide-antibiotic cross-resistance[J]. Microbiol China, 2020, 47(9):2984-2995. |
| [1] | 陈勇, 李亚鑫, 王亚瑄, 梁露洁, 冯思源, 田国宝. MCR-1介导多黏菌素耐药性的分子机制研究进展[J]. 生物技术通报, 2023, 39(6): 102-108. |
| [2] | 任思雨, 姜聪一, 于涛, 康瑞, 姜晓冰. agr系统在单核增生细胞李斯特菌耐药及生物被膜形成中的作用[J]. 生物技术通报, 2023, 39(2): 254-262. |
| [3] | 李海利, 郎利敏, 张青娴, 游一, 朱文豪, 王治方, 张立宪, 王克领. 同时产碳青霉烯酶NDM-1和NDM-5的猪源大肠埃希氏菌的鉴定及耐药性研究[J]. 生物技术通报, 2022, 38(9): 106-115. |
| [4] | 文畅, 刘晨, 卢诗韵, 许忠兵, 艾超凡, 廖汉鹏, 周顺桂. 一株新的多重耐药福氏志贺菌噬菌体生物学特性及基因组分析[J]. 生物技术通报, 2022, 38(9): 127-135. |
| [5] | 鲁兆祥, 王夕冉, 连新磊, 廖晓萍, 刘雅红, 孙坚. 基于功能宏基因组学挖掘抗生素耐药基因研究进展[J]. 生物技术通报, 2022, 38(9): 17-27. |
| [6] | 胡功政, 崔小蝶, 翟亚军, 贺丹丹. 细菌黏菌素耐药性及其逆转机制研究进展[J]. 生物技术通报, 2022, 38(9): 28-34. |
| [7] | 刘艺云, 邓利敏, 岳慧颖, 岳超, 刘健华. 质粒接合转移及其抑制剂的研究进展[J]. 生物技术通报, 2022, 38(9): 35-46. |
| [8] | 刘成程, 胡小芳, 冯友军. 细菌耐药:生化机制与应对策略[J]. 生物技术通报, 2022, 38(9): 4-16. |
| [9] | 赵艳坤, 刘慧敏, 孟璐, 王成, 王加启, 郑楠. 大肠埃希菌异质性耐药的研究进展[J]. 生物技术通报, 2022, 38(9): 59-71. |
| [10] | 刘晓黎, 童真艺, 赵亮, 尹丽, 刘晨光. 非抗生素类活性物质抗幽门螺杆菌研究进展[J]. 生物技术通报, 2022, 38(9): 96-105. |
| [11] | 刘警鞠, 张雨森, 陈娟, 孙炳达, 赵国柱. 曲霉属的现代分类命名研究进展[J]. 生物技术通报, 2022, 38(7): 109-118. |
| [12] | 陈福暖, 黄瑜, 蔡佳, 王忠良, 简纪常, 王蓓. ABC转运蛋白结构及其在细菌致病性中的研究进展[J]. 生物技术通报, 2022, 38(6): 43-52. |
| [13] | 赵海晴, 李耘, 梁严内, 刘哲, 任亚林, 李金娟. 联合用药对嗜水气单胞菌耐药性影响研究进展[J]. 生物技术通报, 2022, 38(6): 53-65. |
| [14] | 朱浩, 张严伟, 刘润, 梁艳, 杨奕, 徐天乐, 杨章平. 抗生素佐剂与抗生素联用的抑菌作用研究进展[J]. 生物技术通报, 2022, 38(6): 66-73. |
| [15] | 王楠, 苏誉, 刘文杰, 封明, 毛瑜, 张新国. 植物内生菌中抗耐药微生物活性成分的研究进展[J]. 生物技术通报, 2021, 37(8): 263-274. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||