








生物技术通报 ›› 2022, Vol. 38 ›› Issue (12): 184-193.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0404
张业猛1,2,3( ), 朱丽丽1,2,3, 陈志国2,3(
), 朱丽丽1,2,3, 陈志国2,3( )
)
收稿日期:2022-03-31
出版日期:2022-12-26
发布日期:2022-12-29
作者简介:张业猛,男,博士研究生,研究方向:作物遗传育种;E-mail:基金资助:
ZHANG Ye-meng1,2,3( ), ZHU Li-li1,2,3, CHEN Zhi-guo2,3(
), ZHU Li-li1,2,3, CHEN Zhi-guo2,3( )
)
Received:2022-03-31
Published:2022-12-26
Online:2022-12-29
摘要:
探讨藜麦NHX基因家族生物学特性及盐胁迫下的表达模式,以期为藜麦抗盐品种选择提供理论依据。以藜麦含有Na+/H+ exchange结构域的蛋白质序列为基础,对其基因结构、蛋白质motif、理化性质以及盐胁迫下转录表达进行分析。结果表明,藜麦基因组中含有46个NHXs基因家族成员,氨基酸长度为109-1 820 aa,分子质量为11 472.74-204 381.38 Da,等电点为5.00-9.27,亲水性为-0.226-1.112,不稳定指数为21.71-50.73。进化树分析可以将其分为5个亚组,在进化树中位置相近的NHXs基因具有相近的结构。同一亚组的NHXs蛋白具有相似motif结构,与拟南芥、小麦和玉米为直系同源蛋白,且多数定位在细胞膜和液泡中。通过盐胁迫下藜麦转录组数据分析发现,随盐胁迫时间增加,其中有26个NHXs基因的转录水平提升,9个下降,11个无明显变化。此外,35个受到盐胁迫诱导表达的NHXs基因,通过GO注释发现,主要参与离子、细胞稳态和离子跨膜运输过程。NHXs基因结构和蛋白质motif的多样性,推测不同亚组NHXs在盐胁迫中参与不同的响应过程,其盐胁迫下不同的表达模式也表明在盐胁迫响应过程中发挥重要作用。
张业猛, 朱丽丽, 陈志国. 藜麦NHX基因家族鉴定及盐胁迫下表达分析[J]. 生物技术通报, 2022, 38(12): 184-193.
ZHANG Ye-meng, ZHU Li-li, CHEN Zhi-guo. Identification and Expression Analysis of NHX Gene Family in Quinoa Under Salt Stress[J]. Biotechnology Bulletin, 2022, 38(12): 184-193.
| 基因Gene | 上游引物Forward primer(5'-3') | 下游引物Reverse primer(5'-3') |
|---|---|---|
| AUR62041754 | ATGGAGGAGGGGAAGGGA | TTAAGAGGTACCCTTTTTTTCCT |
| AUR62027477 | ATGTTCGATCGTGCTTTGGAG | TCATGAATCATGAGACCCCCCTT |
| AUR62043892 | ATGGACCTTTCTGTGGTTGGC | CTAGTAACAAGCCCAAGGAAC |
| AUR62017691 | ATGGAGGATCAGCAGATTTCTCC | TCAGTTACGAGTACTGTAGACTT |
| ACT1[ | GTCCACAGGTGCTTCTAAG | AACAACTCCTCACCTTCTCATG |
表1 藜麦NHXs基因实时荧光定量引物
Table 1 RT-qPCR primers of NHXs in quinoa
| 基因Gene | 上游引物Forward primer(5'-3') | 下游引物Reverse primer(5'-3') |
|---|---|---|
| AUR62041754 | ATGGAGGAGGGGAAGGGA | TTAAGAGGTACCCTTTTTTTCCT |
| AUR62027477 | ATGTTCGATCGTGCTTTGGAG | TCATGAATCATGAGACCCCCCTT |
| AUR62043892 | ATGGACCTTTCTGTGGTTGGC | CTAGTAACAAGCCCAAGGAAC |
| AUR62017691 | ATGGAGGATCAGCAGATTTCTCC | TCAGTTACGAGTACTGTAGACTT |
| ACT1[ | GTCCACAGGTGCTTCTAAG | AACAACTCCTCACCTTCTCATG |
| 基因 ID Gene ID | 氨基酸 Number of amino acids/aa | 分子质量 Molecular weight/Da | 等电点 pI | 亲水性 Gravy | 不稳定指数 Instability index | 亚细胞预测 Predicted location |
|---|---|---|---|---|---|---|
| AUR62017691 | 514 | 57 092.24 | 5.38 | 0.439 | 50.73 | 液泡Vacuole |
| AUR62044447 | 618 | 69 784.72 | 5.54 | 0.373 | 32.07 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62023671 | 510 | 55 482.83 | 7.06 | 0.747 | 26.34 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62022028 | 618 | 69 784.72 | 5.54 | 0.373 | 32.07 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62020843- | 517 | 58 012.47 | 8.38 | 0.460 | 32.20 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62017877 | 109 | 11 472.74 | 6.74 | 1.112 | 21.71 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62021694 | 665 | 74 913.30 | 6.68 | 0.403 | 30.77 | 液泡 Vacuole |
| AUR62019781 | 804 | 86 860.99 | 8.88 | 0.360 | 40.84 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62021703 | 607 | 68 131.50 | 5.57 | 0.432 | 47.58 | 液泡 Vacuole |
| AUR62008394 | 598 | 66 617.46 | 5.97 | 0.352 | 33.74 | 液泡 Vacuole |
| AUR62021702 | 576 | 64 055.50 | 5.94 | 0.387 | 35.34 | 液泡 Vacuole |
| AUR62022953 | 713 | 79 785.44 | 8.69 | 0.291 | 34.83 | 液泡 Vacuole |
| AUR62009662 | 684 | 75 352.40 | 6.43 | 0.385 | 37.95 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62004319 | 1 227 | 131 988.87 | 5.08 | 0.050 | 41.71 | 细胞核 Nucleus |
| AUR62027477 | 486 | 52 731.20 | 7.08 | 0.696 | 32.95 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62019779 | 801 | 86 091.87 | 8.44 | 0.360 | 36.40 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62043892 | 676 | 76 288.29 | 7.91 | 0.300 | 32.05 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62008393 | 607 | 67 909.32 | 5.65 | 0.440 | 47.78 | 液泡 Vacuole |
| AUR62009709 | 867 | 96 471.46 | 9.23 | 0.297 | 46.97 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62033334 | 1 214 | 130 455.31 | 5.09 | 0.084 | 39.66 | 细胞核 Nucleus |
| AUR62041754 | 349 | 36 309.11 | 8.89 | 0.781 | 25.43 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62033325 | 1 181 | 126 888.57 | 5.16 | 0.113 | 39.66 | 细胞核 Nucleus |
| AUR62001874 | 429 | 47 260.98 | 6.45 | 0.636 | 33.44 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62001864 | 753 | 83 016.74 | 7.88 | 0.396 | 38.15 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62009707 | 704 | 79 132.21 | 8.20 | 0.242 | 41.03 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62000934 | 551 | 60 928.18 | 6.49 | 0.534 | 34.53 | 液泡 Vacuole |
| AUR62027069 | 791 | 85 976.49 | 5.20 | 0.320 | 34.07 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62019780 | 801 | 86 024.90 | 8.13 | 0.387 | 40.33 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62036238 | 759 | 85 128.10 | 8.39 | 0.229 | 38.77 | 液泡 Vacuole |
| AUR62018573 | 779 | 86 152.60 | 8.41 | 0.299 | 31.27 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62026204 | 778 | 86 638.91 | 7.90 | 0.174 | 38.40 | 液泡 Vacuole |
| AUR62009395 | 1 646 | 182 928.70 | 8.87 | -0.226 | 39.57 | 液泡 Vacuole |
| AUR62008401 | 665 | 74 930.13 | 6.52 | 0.407 | 31.41 | 液泡 Vacuole |
| AUR62009673 | 1 820 | 204 381.58 | 6.58 | 0.332 | 37.00 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62001875 | 1 340 | 150 822.00 | 6.78 | 0.353 | 33.17 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62001876 | 674 | 75 685.34 | 5.54 | 0.340 | 39.06 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62022963 | 492 | 53 620.84 | 9.00 | 0.703 | 40.62 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62000862 | 544 | 60 147.10 | 6.08 | 0.470 | 32.40 | 液泡 Vacuole |
| AUR62021295 | 548 | 59 615.20 | 5.58 | 0.675 | 25.39 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62030239 | 527 | 59 096.47 | 5.00 | 0.185 | 42.68 | 液泡 Vacuole |
| AUR62003573 | 797 | 86 437.67 | 6.33 | 0.318 | 38.20 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62036236 | 844 | 93 703.46 | 9.27 | 0.309 | 48.18 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62003491 | 1 211 | 133 817.30 | 6.26 | 0.088 | 35.14 | 细胞膜 Cell membrane |
| AUR62009672 | 648 | 71 771.52 | 5.28 | 0.331 | 33.94 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62026390 | 759 | 84 042.11 | 6.83 | 0.273 | 37.52 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62033999 | 491 | 53 035.55 | 7.08 | 0.709 | 31.97 | 细胞膜、液泡Cell membrane,vacuole |
表2 藜麦NHXs蛋白的理化性质及亚细胞定位
Table 2 Physicochemical properties and subcellular localizations of quinoa NHXs proteins
| 基因 ID Gene ID | 氨基酸 Number of amino acids/aa | 分子质量 Molecular weight/Da | 等电点 pI | 亲水性 Gravy | 不稳定指数 Instability index | 亚细胞预测 Predicted location |
|---|---|---|---|---|---|---|
| AUR62017691 | 514 | 57 092.24 | 5.38 | 0.439 | 50.73 | 液泡Vacuole |
| AUR62044447 | 618 | 69 784.72 | 5.54 | 0.373 | 32.07 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62023671 | 510 | 55 482.83 | 7.06 | 0.747 | 26.34 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62022028 | 618 | 69 784.72 | 5.54 | 0.373 | 32.07 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62020843- | 517 | 58 012.47 | 8.38 | 0.460 | 32.20 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62017877 | 109 | 11 472.74 | 6.74 | 1.112 | 21.71 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62021694 | 665 | 74 913.30 | 6.68 | 0.403 | 30.77 | 液泡 Vacuole |
| AUR62019781 | 804 | 86 860.99 | 8.88 | 0.360 | 40.84 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62021703 | 607 | 68 131.50 | 5.57 | 0.432 | 47.58 | 液泡 Vacuole |
| AUR62008394 | 598 | 66 617.46 | 5.97 | 0.352 | 33.74 | 液泡 Vacuole |
| AUR62021702 | 576 | 64 055.50 | 5.94 | 0.387 | 35.34 | 液泡 Vacuole |
| AUR62022953 | 713 | 79 785.44 | 8.69 | 0.291 | 34.83 | 液泡 Vacuole |
| AUR62009662 | 684 | 75 352.40 | 6.43 | 0.385 | 37.95 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62004319 | 1 227 | 131 988.87 | 5.08 | 0.050 | 41.71 | 细胞核 Nucleus |
| AUR62027477 | 486 | 52 731.20 | 7.08 | 0.696 | 32.95 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62019779 | 801 | 86 091.87 | 8.44 | 0.360 | 36.40 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62043892 | 676 | 76 288.29 | 7.91 | 0.300 | 32.05 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62008393 | 607 | 67 909.32 | 5.65 | 0.440 | 47.78 | 液泡 Vacuole |
| AUR62009709 | 867 | 96 471.46 | 9.23 | 0.297 | 46.97 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62033334 | 1 214 | 130 455.31 | 5.09 | 0.084 | 39.66 | 细胞核 Nucleus |
| AUR62041754 | 349 | 36 309.11 | 8.89 | 0.781 | 25.43 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62033325 | 1 181 | 126 888.57 | 5.16 | 0.113 | 39.66 | 细胞核 Nucleus |
| AUR62001874 | 429 | 47 260.98 | 6.45 | 0.636 | 33.44 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62001864 | 753 | 83 016.74 | 7.88 | 0.396 | 38.15 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62009707 | 704 | 79 132.21 | 8.20 | 0.242 | 41.03 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62000934 | 551 | 60 928.18 | 6.49 | 0.534 | 34.53 | 液泡 Vacuole |
| AUR62027069 | 791 | 85 976.49 | 5.20 | 0.320 | 34.07 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62019780 | 801 | 86 024.90 | 8.13 | 0.387 | 40.33 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62036238 | 759 | 85 128.10 | 8.39 | 0.229 | 38.77 | 液泡 Vacuole |
| AUR62018573 | 779 | 86 152.60 | 8.41 | 0.299 | 31.27 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62026204 | 778 | 86 638.91 | 7.90 | 0.174 | 38.40 | 液泡 Vacuole |
| AUR62009395 | 1 646 | 182 928.70 | 8.87 | -0.226 | 39.57 | 液泡 Vacuole |
| AUR62008401 | 665 | 74 930.13 | 6.52 | 0.407 | 31.41 | 液泡 Vacuole |
| AUR62009673 | 1 820 | 204 381.58 | 6.58 | 0.332 | 37.00 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62001875 | 1 340 | 150 822.00 | 6.78 | 0.353 | 33.17 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62001876 | 674 | 75 685.34 | 5.54 | 0.340 | 39.06 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62022963 | 492 | 53 620.84 | 9.00 | 0.703 | 40.62 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62000862 | 544 | 60 147.10 | 6.08 | 0.470 | 32.40 | 液泡 Vacuole |
| AUR62021295 | 548 | 59 615.20 | 5.58 | 0.675 | 25.39 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62030239 | 527 | 59 096.47 | 5.00 | 0.185 | 42.68 | 液泡 Vacuole |
| AUR62003573 | 797 | 86 437.67 | 6.33 | 0.318 | 38.20 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62036236 | 844 | 93 703.46 | 9.27 | 0.309 | 48.18 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62003491 | 1 211 | 133 817.30 | 6.26 | 0.088 | 35.14 | 细胞膜 Cell membrane |
| AUR62009672 | 648 | 71 771.52 | 5.28 | 0.331 | 33.94 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62026390 | 759 | 84 042.11 | 6.83 | 0.273 | 37.52 | 细胞膜、液泡Cell membrane,vacuole |
| AUR62033999 | 491 | 53 035.55 | 7.08 | 0.709 | 31.97 | 细胞膜、液泡Cell membrane,vacuole |

图 4 拟南芥、小麦、玉米和藜麦的NHXs蛋白的系统进化分析 AT:拟南芥;Tr:小麦;Zm:玉米;AU:藜麦
Fig. 4 Phylogenetic analysis of NHXs proteins in Arabidopsis,wheat,maize and quinoa AT:Arabidopsis thaliana;Tr:Triticum aestivum L.;Zm:Zea mays;AU:Chenopodium quinoa Willd.
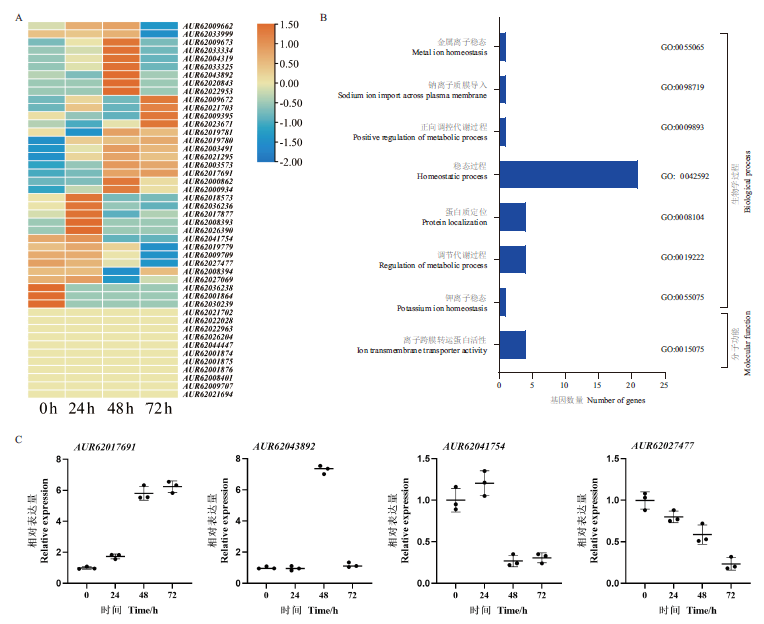
图5 藜麦NHXs基因表达模式与GO注释 A:NaCl胁迫下NHX基因家族表达模式;B:NHX基因家族GO注释;C:NHXs基因RT-qPCR分析
Fig. 5 Gene expression patterns and GO annotations of NHXs in quinoa A:Expression pattern analysis of NHX family under NaCl stress. B:GO annotation of NHX gene family. C:RT-qPCR analysis of NHXs genes
| [1] |
朱健康, 倪建平. 植物非生物胁迫信号转导及应答[J]. 中国稻米, 2016, 22(6):52-60.
doi: 10.3969/j.issn.1006-8082.2016.06.012 |
|
Zhu JK, Ni JP. Abiotic stress signaling and responses in plants[J]. China Rice, 2016, 22(6):52-60.
doi: 10.3969/j.issn.1006-8082.2016.06.012 |
|
| [2] |
Munns R, Tester M. Mechanisms of salinity tolerance[J]. Annu Rev Plant Biol, 2008, 59:651-681.
doi: 10.1146/annurev.arplant.59.032607.092911 pmid: 18444910 |
| [3] |
Hanin M, Brini F, Ebel C, et al. Plant dehydrins and stress tolerance:versatile proteins for complex mechanisms[J]. Plant Signal Behav, 2011, 6(10):1503-1509.
doi: 10.4161/psb.6.10.17088 pmid: 21897131 |
| [4] |
Waditee R, Hibino T, Tanaka Y, et al. Halotolerant cyanobacterium Aphanothece halophytica contains an Na+/H+ antiporter, homologous to eukaryotic ones, with novel ion specificity affected by C-terminal tail[J]. J Biol Chem, 2001, 276(40):36931-36938.
doi: 10.1074/jbc.M103650200 pmid: 11479290 |
| [5] | Cao D, Hou WS, Liu W, et al. Overexpression of TaNHX2 enhances salt tolerance of ‘composite’and whole transgenic soybean plants[J]. Plant Cell Tissue Organ Cult PCTOC, 2011, 107(3):541-552. |
| [6] |
Ratner A, Jacoby B. Effect of K+, its counter anion, and pH on sodium efflux from barley root tips[J]. J Exp Bot, 1976, 27(5):843-852.
doi: 10.1093/jxb/27.5.843 URL |
| [7] |
Gaxiola RA, Rao R, Sherman A, et al. The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast[J]. Proc Natl Acad Sci USA, 1999, 96(4):1480-1485.
doi: 10.1073/pnas.96.4.1480 URL |
| [8] |
Fukuda A, Nakamura A, Tagiri A, et al. Function, intracellular localization and the importance in salt tolerance of a vacuolar Na+/H+ antiporter from rice[J]. Plant Cell Physiol, 2004, 45(2):146-159.
doi: 10.1093/pcp/pch014 URL |
| [9] |
Brini F, Gaxiola RA, Berkowitz GA, et al. Cloning and characterization of a wheat vacuolar cation/proton antiporter and pyrophosphatase proton pump[J]. Plant Physiol Biochem, 2005, 43(4):347-354.
doi: 10.1016/j.plaphy.2005.02.010 URL |
| [10] |
Huang Y, Zhang XX, Li YH, et al. Overexpression of the Suaeda salsa SsNHX1 gene confers enhanced salt and drought tolerance to transgenic Zea mays[J]. J Integr Agric, 2018, 17(12):2612-2623.
doi: 10.1016/S2095-3119(18)61998-7 URL |
| [11] |
Barragán V, Leidi EO, Andrés Z, et al. Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis[J]. Plant Cell, 2012, 24(3):1127-1142.
doi: 10.1105/tpc.111.095273 URL |
| [12] |
Qiu QS. Plant and yeast NHX antiporters:roles in membrane trafficking[J]. J Integr Plant Biol, 2012, 54(2):66-72.
doi: 10.1111/j.1744-7909.2012.01097.x URL |
| [13] |
Dragwidge JM, Ford BA, Ashnest JR, et al. Two endosomal NHX-type Na+/H+ antiporters are involved in auxin-mediated development in Arabidopsis thaliana[J]. Plant Cell Physiol, 2018, 59(8):1660-1669.
doi: 10.1093/pcp/pcy090 pmid: 29788486 |
| [14] |
Wang J, Qiu NW, Wang P, et al. Na+ compartmentation strategy of Chinese cabbage in response to salt stress[J]. Plant Physiol Biochem, 2019, 140:151-157.
doi: 10.1016/j.plaphy.2019.05.001 URL |
| [15] |
Li WYF, Wong FL, Tsai SN, et al. Tonoplast-located GmCLC1 and GmNHX1 from soybean enhance NaCl tolerance in transgenic bright yellow(BY)-2 cells[J]. Plant Cell Environ, 2006, 29(6):1122-1137.
doi: 10.1111/j.1365-3040.2005.01487.x URL |
| [16] |
Ayadi M, Martins V, Ben Ayed R, et al. Genome wide identification, molecular characterization, and gene expression analyses of grapevine NHX antiporters suggest their involvement in growth, ripening, seed dormancy, and stress response[J]. Biochem Genet, 2020, 58(1):102-128.
doi: 10.1007/s10528-019-09930-4 pmid: 31286319 |
| [17] | 董丽君, 刘灵娣, 车文利, 等. 转Na+/H+逆向转运蛋白基因(AlNHX1)大豆纯合系筛选及生理分析[J]. 大豆科学, 2015, 34(3):378-383. |
| Dong LJ, Liu LD, Che WL, et al. Screening of homogeneous transgenic lines with Na+/H+ antiporter gene(AlNHX1)in soybean and its physiological analysis[J]. Soybean Sci, 2015, 34(3):378-383. | |
| [18] | Yarra R, He SJ, Abbagani S, et al. Overexpression of a wheat Na+/H+ antiporter gene(TaNHX2)enhances tolerance to salt stress in transgenic tomato plants(Solanum lycopersicum L.)[J]. Plant Cell Tissue Organ Cult PCTOC, 2012, 111(1):49-57. |
| [19] |
Li WH, Du J, Feng HM, et al. Function of NHX-type transporters in improving rice tolerance to aluminum stress and soil acidity[J]. Planta, 2020, 251(3):71.
doi: 10.1007/s00425-020-03361-x pmid: 32108903 |
| [20] |
Jacobsen SE, Mujica A, Jensen CR. The resistance of quinoa(Chenopodium quinoaWilld. )to adverse abiotic factors[J]. Food Rev Int, 2003, 19(1/2):99-109.
doi: 10.1081/FRI-120018872 URL |
| [21] | González JA, Gallardo M, Hilal M, et al. Physiological responses of quinoa(Chenopodium quinoa Willd. )to drought and waterlogging stresses:dry matter partitioning[J]. Bot Stud, 2009, 50(1):35-42. |
| [22] |
Adolf VI, Shabala S, Andersen MN, et al. Varietal differences of quinoa’s tolerance to saline conditions[J]. Plant Soil, 2012, 357(1/2):117-129.
doi: 10.1007/s11104-012-1133-7 URL |
| [23] |
Jarvis DE, Ho YS, Lightfoot DJ, et al. The genome of Chenopodium quinoa[J]. Nature, 2017, 542(7641):307-312.
doi: 10.1038/nature21370 URL |
| [24] | 贾冰晨, 王宇, 张东亮, 等. 藜麦内参基因筛选及盐胁迫相关基因表达分析[J]. 烟台大学学报:自然科学与工程版, 2020, 33(3):283-288. |
| Jia BC, Wang Y, Zhang DL, et al. Screening of reference genes and analysis of gene expression under salt stress in Chenopodium quinoa[J]. J Yantai Univ Nat Sci Eng Ed, 2020, 33(3):283-288. | |
| [25] |
Ohnishi M, Fukada-Tanaka S, Hoshino A, et al. Characterization of a novel Na+/H+ antiporter gene InNHX2 and comparison of InNHX2 with InNHX1, which is responsible for blue flower coloration by increasing the vacuolar pH in the Japanese morning glory[J]. Plant Cell Physiol, 2005, 46(2):259-267.
pmid: 15695437 |
| [26] | 卢世雄, 许春苗, 何红红, 等. 葡萄NHX基因家族的鉴定和表达分析[J]. 果树学报, 2019, 36(3):266-276. |
| Lu SX, Xu CM, He HH, et al. Identification and expression analysis of NHX genes family in grape[J]. J Fruit Sci, 2019, 36(3):266-276. | |
| [27] | 徐亚, 滕梦鑫, 何岳东, 等. 香蕉NHX基因家族的鉴定及表达分析[J]. 植物生理学报, 2021, 57(3):681-691. |
|
Xu Y, Teng MX, He YD, et al. Identification and expression analysis of NHX genes family in banana[J]. Plant Physiol J, 2021, 57(3):681-691.
doi: 10.1104/pp.57.5.681 URL |
|
| [28] | 李静, 刘明, 孙晶, 等. Na+(K+)/H+转运蛋白NHX基因的研究进展[J]. 大豆科学, 2011, 30(6):1035-1039. |
| Li J, Liu M, Sun J, et al. Progress of research on Na+(K+)/H+ antiporter NHX gene[J]. Soybean Sci, 2011, 30(6):1035-1039. | |
| [29] |
Lu SY, Jing YX, Shen SH, et al. Antiporter gene from Hordum brevisubulatum(Trin. )link and its overexpression in transgenic tobaccos[J]. J Integr Plant Biol, 2005, 47(3):343-349.
doi: 10.1111/j.1744-7909.2005.00027.x URL |
| [30] | 杨杰, 陈蓉, 胡文娟, 等. 甜橙NHX基因家族的鉴定及功能分析[J]. 江苏农业科学, 2022, 50(7):35-42. |
| Yang J, Chen R, Hu WJ, et al. Identification and functional analysis of NHX gene family in Citrus sinensis[J]. Jiangsu Agric Sci, 2022, 50(7):35-42. | |
| [31] | 郝东风, 兰海燕, 陈邦党, 等. 新疆盐生植物耐盐基因NHX转化甘蓝型油菜及其耐盐性的初步研究[J]. 生物技术通报, 2006(3):81-84. |
| Hao DF, Lan HY, Chen BD, et al. Introduction of Na+/H+ transporter gene NHX from native Halophyte salicornia spp in Xinjiang into Brassica napus and its salt tolerance[J]. Biotechnol Bull, 2006(3):81-84. | |
| [32] | 赵云霞, 郭丹丽, 魏艳玲, 等. 新疆无苞芥Na+/H+逆向转运蛋白基因OpNHX1的克隆、表达分析与功能验证[J]. 生物技术通报, 2014(7):74-80. |
| Zhao YX, Guo DL, Wei YL, et al. Cloning, expressing and functional analysis of Na+/H+ antiporter gene OpNHX1 from Olimarabidop-sis pumila in Xinjiang[J]. Biotechnol Bull, 2014(7):74-80. | |
| [33] |
Ruiz KB, Maldonado J, Biondi S, et al. RNA-seq analysis of salt-stressed versus non salt-stressed transcriptomes of Chenopodium quinoa Landrace R49[J]. Genes, 2019, 10(12):1042.
doi: 10.3390/genes10121042 URL |
| [1] | 王帅, 冯宇梅, 白苗, 杜维俊, 岳爱琴. 大豆GmHMGR基因响应外源激素及非生物胁迫功能研究[J]. 生物技术通报, 2023, 39(7): 131-142. |
| [2] | 魏茜雅, 秦中维, 梁腊梅, 林欣琪, 李映志. 褪黑素种子引发处理提高朝天椒耐盐性的作用机制[J]. 生物技术通报, 2023, 39(7): 160-172. |
| [3] | 张路阳, 韩文龙, 徐晓雯, 姚健, 李芳芳, 田效园, 张智强. 烟草TCP基因家族的鉴定及表达分析[J]. 生物技术通报, 2023, 39(6): 248-258. |
| [4] | 王海龙, 李雨倩, 王勃, 邢国芳, 张杰伟. 谷子SiMAPK3基因的克隆和表达特性分析[J]. 生物技术通报, 2023, 39(3): 123-132. |
| [5] | 杜清洁, 周璐瑶, 杨思震, 张嘉欣, 陈春林, 李娟起, 李猛, 赵士文, 肖怀娟, 王吉庆. 过表达CaCP1提高转基因烟草对盐胁迫的敏感性[J]. 生物技术通报, 2023, 39(2): 172-182. |
| [6] | 汪明滔, 刘建伟, 赵春钊. 植物调控盐胁迫下细胞壁完整性的分子机制[J]. 生物技术通报, 2023, 39(11): 18-27. |
| [7] | 张玉娟, 黎冬华, 宫慧慧, 崔新晓, 高春华, 张秀荣, 游均, 赵军胜. 芝麻NAC转录因子基因SiNAC77的克隆及耐盐功能分析[J]. 生物技术通报, 2023, 39(11): 308-317. |
| [8] | 徐扬, 丁红, 张冠初, 郭庆, 张智猛, 戴良香. 盐胁迫下花生种子萌发期代谢组学分析[J]. 生物技术通报, 2023, 39(1): 199-213. |
| [9] | 陈光, 李佳, 杜瑞英, 王旭. 水稻盐敏感突变体ss2的鉴定与基因功能分析[J]. 生物技术通报, 2022, 38(9): 158-166. |
| [10] | 赵林艳, 官会林, 王克书, 卢燕磊, 向萍, 魏富刚, 杨绍周, 徐武美. 土壤含水量对三七连作土壤微生物群落的影响[J]. 生物技术通报, 2022, 38(7): 215-223. |
| [11] | 赵忠娟, 杨凯, 扈进冬, 魏艳丽, 李玲, 徐维生, 李纪顺. 盐胁迫条件下哈茨木霉ST02对椒样薄荷生长及根区土壤理化性质的影响[J]. 生物技术通报, 2022, 38(7): 224-235. |
| [12] | 杨佳宝, 周至铭, 张展, 冯丽, 孙黎. 向日葵HaLACS1的克隆、表达及酵母功能互补鉴定[J]. 生物技术通报, 2022, 38(6): 147-156. |
| [13] | 张斌, 杨昕霞. 水稻响应盐胁迫关键转录因子的鉴定[J]. 生物技术通报, 2022, 38(3): 9-15. |
| [14] | 曹映辉, 胡美娟, 童妍, 张燕萍, 赵凯, 彭东辉, 周育真. 建兰ABC基因家族鉴定及其在花发育过程中的表达模式分析[J]. 生物技术通报, 2022, 38(11): 162-174. |
| [15] | 张彤彤, 郑登俞, 吴忠义, 张中保, 于荣. 玉米NF-Y转录因子基因ZmNF-YB13响应干旱和盐胁迫的功能分析[J]. 生物技术通报, 2022, 38(10): 115-123. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||