








生物技术通报 ›› 2023, Vol. 39 ›› Issue (2): 126-138.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0567
穆德添1( ), 万凌云2, 章瑶1, 韦树根2, 陆英1, 付金娥2, 田艺1, 潘丽梅2(
), 万凌云2, 章瑶1, 韦树根2, 陆英1, 付金娥2, 田艺1, 潘丽梅2( ), 唐其1(
), 唐其1( )
)
收稿日期:2022-05-09
出版日期:2023-02-26
发布日期:2023-03-07
作者简介:穆德添,男,硕士研究生,研究方向:药用植物分子生物学;E-mail: 基金资助:
MU De-tian1( ), WAN Ling-yun2, ZHANG Yao1, WEI Shu-gen2, LU Ying1, FU Jin-e2, TIAN Yi1, PAN Li-mei2(
), WAN Ling-yun2, ZHANG Yao1, WEI Shu-gen2, LU Ying1, FU Jin-e2, TIAN Yi1, PAN Li-mei2( ), TANG Qi1(
), TANG Qi1( )
)
Received:2022-05-09
Published:2023-02-26
Online:2023-03-07
摘要:
为研究钩藤不同部位中最适管家基因与钩藤生物碱上游合成途径中关键酶基因的表达模式。以钩藤根、茎钩、叶片、蒴果为实验材料,通过RT-qPCR技术、GeNorm、NormFinder、Bestkeeper软件及△Ct程序、RefFinder在线网站分析了12个候选管家基因(18S、SAM、TUA、TUB、EF-1β、EF-1α、RNA L13、GAPDH、Actin6、PAL、CYP、cdc73)表达稳定性。结果表明,最适管家基因为SAM。再以SAM为管家基因,基于“基因组+转录组+代谢组”共表达分析筛选出钩藤生物碱上游合成途径中的15个重点候选相关基因(G8H、8-HGO、IS、CYP76A26、7-DLGT、7-DLH、LAMT、SLS、AS、AnPRT、IGPS、TSA、TSB、TDC、STR)进行表达分析,结果显示这些基因的表达量与含量趋势较为一致,很可能参与钩藤中TIAs的合成。
穆德添, 万凌云, 章瑶, 韦树根, 陆英, 付金娥, 田艺, 潘丽梅, 唐其. 钩藤管家基因筛选及生物碱合成相关基因的表达分析[J]. 生物技术通报, 2023, 39(2): 126-138.
MU De-tian, WAN Ling-yun, ZHANG Yao, WEI Shu-gen, LU Ying, FU Jin-e, TIAN Yi, PAN Li-mei, TANG Qi. House-keeping Genes Screening and Expression Patterns Analysis of Genes Involved in Alkaloid Biosynthesis in Uncaria rhynchophylla[J]. Biotechnology Bulletin, 2023, 39(2): 126-138.
| 基因Gene | 基因全称Gene name | 引物序列Primer sequence(5'-3') |
|---|---|---|
| G8H | Geraniol 8-hydroxylase | F:CGCCCATAATCATTCCACTA R:GCCAATGTCACTGCCTCAC |
| 8-HGO | 8-hydroxygeraniol oxidoreductase | F:GCGTCTGGTGTTCTATCTCCGTT R:GGTTGCGTTGCTGCCTACAT |
| IS | Iridoid synthase | F:ACCCTTCAGAAACTCAACAAAAGC R:CCTCGCAACATTCTGGGCTA |
| CYP76A26 | Nepetalactol monooxygenase | F:ACCTTATGCTGTCGAGGGATT R:TAACACTGCCAGCTTTCCTGT |
| 7-DLGT / UGT85A24 | 7-deoxyloganetic acid glucosyltransferase | F:CATGGAAGAAGACAGCAGGAG R:CATTGTCGAATTCCAACCACT |
| 7-DLH / CYP2A224 | 7-deoxyloganate 7-hydroxylase | F:AGGAACAACAGGATGAGAGCA R:AGGTAGTGTCCTGTCCAGCAA |
| LAMT | Loganate methyltransferase | F:TGTCAAAGAAATGCCTGAAGAAG R:ATGCGGAAAGGCTTAGATGG |
| SLS / CYP72A1 | Secologanin synthase | F:TCGGACCATTCAAACCTACGG R:GCTTGGACCATCTGTCACCCTC |
| AS | Anthranilate synthase | F:TCAAGGACGAAGGGTGGAACAG R:ACCAACCCAACCACCACAAAAT |
| AnPRT | Anthranilate phosphoribosyltransferase | F:GTTGGGACTGGTGGTGATGG R:CTTGACCTATTTCCTTGCTTTGC |
| IGPS | Indole-3-glycerol phosphate synthase | F:GTTGGGGAGTCTGGGCTTTT R:GAGTCCGGTGATTGCCTTAGTT |
| TSA | Tryptophan synthase alpha chain | F:TGTGAAACAAGTTGCTGGATGG R:AGGAGATTTTGCCTCGCCTA |
| TSB | Tryptophan synthase beta chain | F:GCTGAGGTTAGGCCAGTTCATT R:TCCACATTAGTCACCCAGTCCC |
| TDC | L-tryptophan decarboxylase | F:GGCAGGTATTTTCCCACGCA R:GTTCCCACGGTAGCACAGA |
| STR | Strictosidine synthase | F:GCCGATGGTCGGATTCTCAA R:GGCCCAAATAGGCATCAGCA |
表1 钩藤生物碱上游合成途径中关键酶基因RT-qPCR的引物序列
Table 1 Primer sequences for key genes in U. rhynchophylla alkaloids upstream synthesis pathway for RT-qPCR
| 基因Gene | 基因全称Gene name | 引物序列Primer sequence(5'-3') |
|---|---|---|
| G8H | Geraniol 8-hydroxylase | F:CGCCCATAATCATTCCACTA R:GCCAATGTCACTGCCTCAC |
| 8-HGO | 8-hydroxygeraniol oxidoreductase | F:GCGTCTGGTGTTCTATCTCCGTT R:GGTTGCGTTGCTGCCTACAT |
| IS | Iridoid synthase | F:ACCCTTCAGAAACTCAACAAAAGC R:CCTCGCAACATTCTGGGCTA |
| CYP76A26 | Nepetalactol monooxygenase | F:ACCTTATGCTGTCGAGGGATT R:TAACACTGCCAGCTTTCCTGT |
| 7-DLGT / UGT85A24 | 7-deoxyloganetic acid glucosyltransferase | F:CATGGAAGAAGACAGCAGGAG R:CATTGTCGAATTCCAACCACT |
| 7-DLH / CYP2A224 | 7-deoxyloganate 7-hydroxylase | F:AGGAACAACAGGATGAGAGCA R:AGGTAGTGTCCTGTCCAGCAA |
| LAMT | Loganate methyltransferase | F:TGTCAAAGAAATGCCTGAAGAAG R:ATGCGGAAAGGCTTAGATGG |
| SLS / CYP72A1 | Secologanin synthase | F:TCGGACCATTCAAACCTACGG R:GCTTGGACCATCTGTCACCCTC |
| AS | Anthranilate synthase | F:TCAAGGACGAAGGGTGGAACAG R:ACCAACCCAACCACCACAAAAT |
| AnPRT | Anthranilate phosphoribosyltransferase | F:GTTGGGACTGGTGGTGATGG R:CTTGACCTATTTCCTTGCTTTGC |
| IGPS | Indole-3-glycerol phosphate synthase | F:GTTGGGGAGTCTGGGCTTTT R:GAGTCCGGTGATTGCCTTAGTT |
| TSA | Tryptophan synthase alpha chain | F:TGTGAAACAAGTTGCTGGATGG R:AGGAGATTTTGCCTCGCCTA |
| TSB | Tryptophan synthase beta chain | F:GCTGAGGTTAGGCCAGTTCATT R:TCCACATTAGTCACCCAGTCCC |
| TDC | L-tryptophan decarboxylase | F:GGCAGGTATTTTCCCACGCA R:GTTCCCACGGTAGCACAGA |
| STR | Strictosidine synthase | F:GCCGATGGTCGGATTCTCAA R:GGCCCAAATAGGCATCAGCA |
| 基因Gene | 基因全称Gene name | 引物序列Primer sequence(5'-3') | 斜率k | 扩增效率E/% | 相关系数R2 |
|---|---|---|---|---|---|
| 18S | 18S Ribosomal RNA | F:CTTCGGGATCGGAGTAATGA R:GCGGAGTCCTAGAAGCAACA | -3.14 | 0.97 | 0.999 96 |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | F:CAGGAACCCAGAGGAGATT R:GCATCCTTACTTGGAGCAG | -3.41 | 0.96 | 0.999 78 |
| Actin6 | Actin 6 | F:ACCGAGCGTGGTTATTCTT R:TTCCTGCTGCTTCCATTCC | -3.54 | 0.92 | 0.991 79 |
| EF1-β | Elongation factor 1β | F:AAGGCATCCACCAAGAAGA R:AAGGCAACAATGTCACAGC | -3.49 | 0.93 | 0.998 07 |
| TUB | β-Tubulin | F:GGAAGTAATCTGCGACGAG R:TGAGAACAGCACGAGGGAC | -3.12 | 1.09 | 0.998 71 |
| CYP | Cyclophilin | F:CGAGAAAGGCGTGGGAAAG R:TGAGACCCGTTGGTGTTGG | -3.2 | 1.05 | 0.998 35 |
| EF1-α | Elongation factor 1α | F:GAGCGTGAGCGTGGTATTACTAT R:CCAGTGGTGGAGTCAATGATAA | -3.12 | 1.09 | 0.994 50 |
| PAL | Phenylalanine ammonialyase | F:ATCGCTGAATCCTCCAATA R:CCACCCTACTCCACAATACTT | -3.12 | 1.09 | 0.998 68 |
| RNA L13 | Ribosomal protein L13 | F:CCAGGAGAAGGAATGCGAGG R:GACCGGTTTTTACGGCGATG | -3.67 | 0.87 | 0.991 99 |
| SAM | S-adenosylmethionine decarboxylase | F:CACAATCTGGCATACGAAA R:AACTCACTTGGCTGGAAAC | -3.7 | 0.92 | 0.995 17 |
| cdc73 | Cell division control protein 73 | F:TGGTGGCTGTTTTCGTGTT R:TGATGCCGCTTATTCTTGC | -3.44 | 0.95 | 0.990 33 |
| TUA | α-Tubulin | F:TCCCTTCTTGAGCACACTGAT R:CCATCAAACCTCAAAGACGCA | -3.28 | 1.02 | 0.993 96 |
表2 钩藤12个候选管家基因RT-qPCR的引物序列和扩增系数
Table 2 Primer sequence and amplification parameters for 12 candidate reference genes of U. rhynchophylla in RT-qPCR
| 基因Gene | 基因全称Gene name | 引物序列Primer sequence(5'-3') | 斜率k | 扩增效率E/% | 相关系数R2 |
|---|---|---|---|---|---|
| 18S | 18S Ribosomal RNA | F:CTTCGGGATCGGAGTAATGA R:GCGGAGTCCTAGAAGCAACA | -3.14 | 0.97 | 0.999 96 |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | F:CAGGAACCCAGAGGAGATT R:GCATCCTTACTTGGAGCAG | -3.41 | 0.96 | 0.999 78 |
| Actin6 | Actin 6 | F:ACCGAGCGTGGTTATTCTT R:TTCCTGCTGCTTCCATTCC | -3.54 | 0.92 | 0.991 79 |
| EF1-β | Elongation factor 1β | F:AAGGCATCCACCAAGAAGA R:AAGGCAACAATGTCACAGC | -3.49 | 0.93 | 0.998 07 |
| TUB | β-Tubulin | F:GGAAGTAATCTGCGACGAG R:TGAGAACAGCACGAGGGAC | -3.12 | 1.09 | 0.998 71 |
| CYP | Cyclophilin | F:CGAGAAAGGCGTGGGAAAG R:TGAGACCCGTTGGTGTTGG | -3.2 | 1.05 | 0.998 35 |
| EF1-α | Elongation factor 1α | F:GAGCGTGAGCGTGGTATTACTAT R:CCAGTGGTGGAGTCAATGATAA | -3.12 | 1.09 | 0.994 50 |
| PAL | Phenylalanine ammonialyase | F:ATCGCTGAATCCTCCAATA R:CCACCCTACTCCACAATACTT | -3.12 | 1.09 | 0.998 68 |
| RNA L13 | Ribosomal protein L13 | F:CCAGGAGAAGGAATGCGAGG R:GACCGGTTTTTACGGCGATG | -3.67 | 0.87 | 0.991 99 |
| SAM | S-adenosylmethionine decarboxylase | F:CACAATCTGGCATACGAAA R:AACTCACTTGGCTGGAAAC | -3.7 | 0.92 | 0.995 17 |
| cdc73 | Cell division control protein 73 | F:TGGTGGCTGTTTTCGTGTT R:TGATGCCGCTTATTCTTGC | -3.44 | 0.95 | 0.990 33 |
| TUA | α-Tubulin | F:TCCCTTCTTGAGCACACTGAT R:CCATCAAACCTCAAAGACGCA | -3.28 | 1.02 | 0.993 96 |
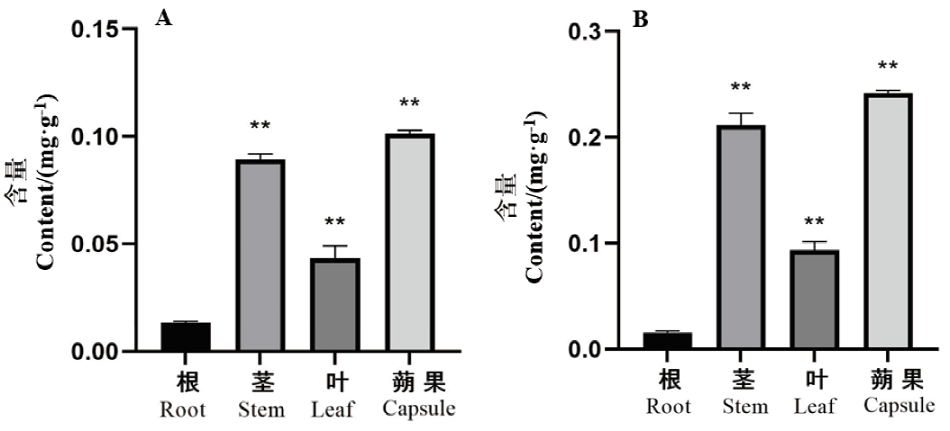
图1 钩藤不同部位中钩藤碱(A)和异钩藤碱(B)的含量
Fig. 1 Rhynchophylline and isorhynchophylline contents from U. rhynchophylla different issues *:P<0.01 ;**:P<0.01, the same below
| 基因 Gene | 几何平均数 GM | 算术平均数 AM | 最小值 Min | 最大值 Max | 标准偏差 Standard deviation | 变异系数 Coefficient of variation | 稳定性排序 Stability rank |
|---|---|---|---|---|---|---|---|
| 18S | 15.55 | 15.55 | 15.09 | 16.15 | 0.32 | 2.05 | 1 |
| SAM | 22.14 | 22.15 | 21.10 | 23.05 | 0.54 | 2.44 | 2 |
| CYP | 19.39 | 19.41 | 18.05 | 20.77 | 0.69 | 3.56 | 3 |
| EF-1β | 21.58 | 21.59 | 20.43 | 22.68 | 0.75 | 3.48 | 4 |
| cdc73 | 24.46 | 24.48 | 23.35 | 26.36 | 0.77 | 3.15 | 5 |
| TUB | 24.30 | 24.31 | 23.42 | 25.68 | 0.78 | 3.22 | 6 |
| EF-1α | 21.05 | 21.07 | 19.34 | 22.51 | 0.87 | 4.12 | 7 |
| PAL | 22.94 | 22.96 | 21.20 | 23.91 | 0.96 | 4.16 | 8 |
| Actin6 | 23.46 | 23.51 | 21.11 | 24.90 | 1.17 | 4.91 | 9 |
| RNA L13 | 22.50 | 22.54 | 21.03 | 25.07 | 1.18 | 5.21 | 10 |
| GAPDH | 26.82 | 26.87 | 24.55 | 28.70 | 1.53 | 5.69 | 11 |
| TUA | 22.36 | 22.53 | 19.15 | 26.03 | 2.78 | 12.34 | 12 |
表3 BestKeeper分析12个候选内参基因表达的稳定性
Table 3 Expression stabilities of 12 candidate reference genes analyzed by BestKeeper
| 基因 Gene | 几何平均数 GM | 算术平均数 AM | 最小值 Min | 最大值 Max | 标准偏差 Standard deviation | 变异系数 Coefficient of variation | 稳定性排序 Stability rank |
|---|---|---|---|---|---|---|---|
| 18S | 15.55 | 15.55 | 15.09 | 16.15 | 0.32 | 2.05 | 1 |
| SAM | 22.14 | 22.15 | 21.10 | 23.05 | 0.54 | 2.44 | 2 |
| CYP | 19.39 | 19.41 | 18.05 | 20.77 | 0.69 | 3.56 | 3 |
| EF-1β | 21.58 | 21.59 | 20.43 | 22.68 | 0.75 | 3.48 | 4 |
| cdc73 | 24.46 | 24.48 | 23.35 | 26.36 | 0.77 | 3.15 | 5 |
| TUB | 24.30 | 24.31 | 23.42 | 25.68 | 0.78 | 3.22 | 6 |
| EF-1α | 21.05 | 21.07 | 19.34 | 22.51 | 0.87 | 4.12 | 7 |
| PAL | 22.94 | 22.96 | 21.20 | 23.91 | 0.96 | 4.16 | 8 |
| Actin6 | 23.46 | 23.51 | 21.11 | 24.90 | 1.17 | 4.91 | 9 |
| RNA L13 | 22.50 | 22.54 | 21.03 | 25.07 | 1.18 | 5.21 | 10 |
| GAPDH | 26.82 | 26.87 | 24.55 | 28.70 | 1.53 | 5.69 | 11 |
| TUA | 22.36 | 22.53 | 19.15 | 26.03 | 2.78 | 12.34 | 12 |
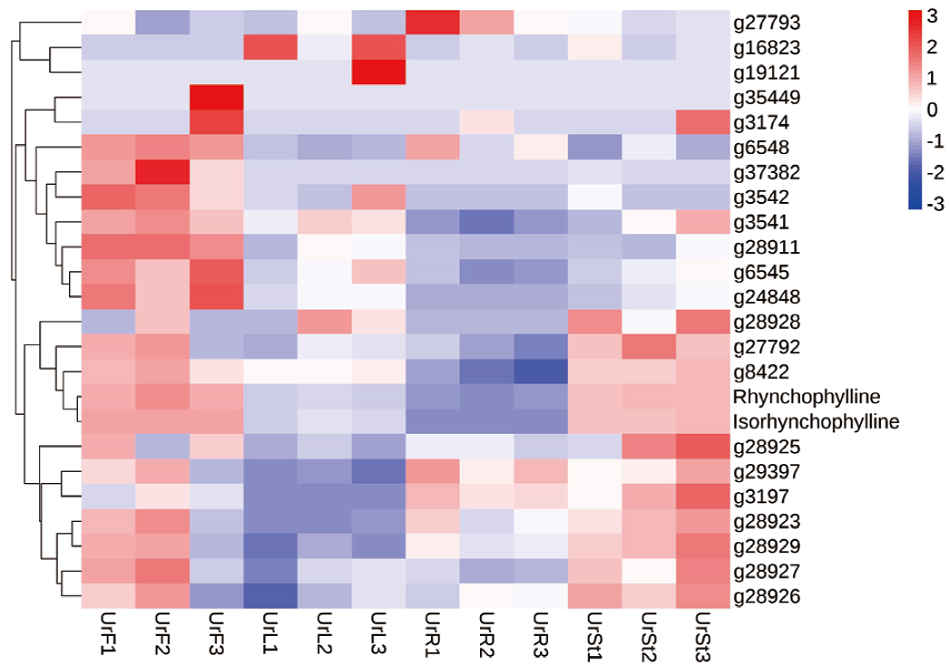
图9 异胡豆苷合酶基因筛选热图 Rhynchophylline为钩藤碱含量,Isorhynchophylline为异钩藤碱含量
Fig. 9 Screening heatmap of strictosidine synthase gene Rhynchophylline is the content of rhynchophylline, and isorhynchophylline is the content of isorhynchophylline
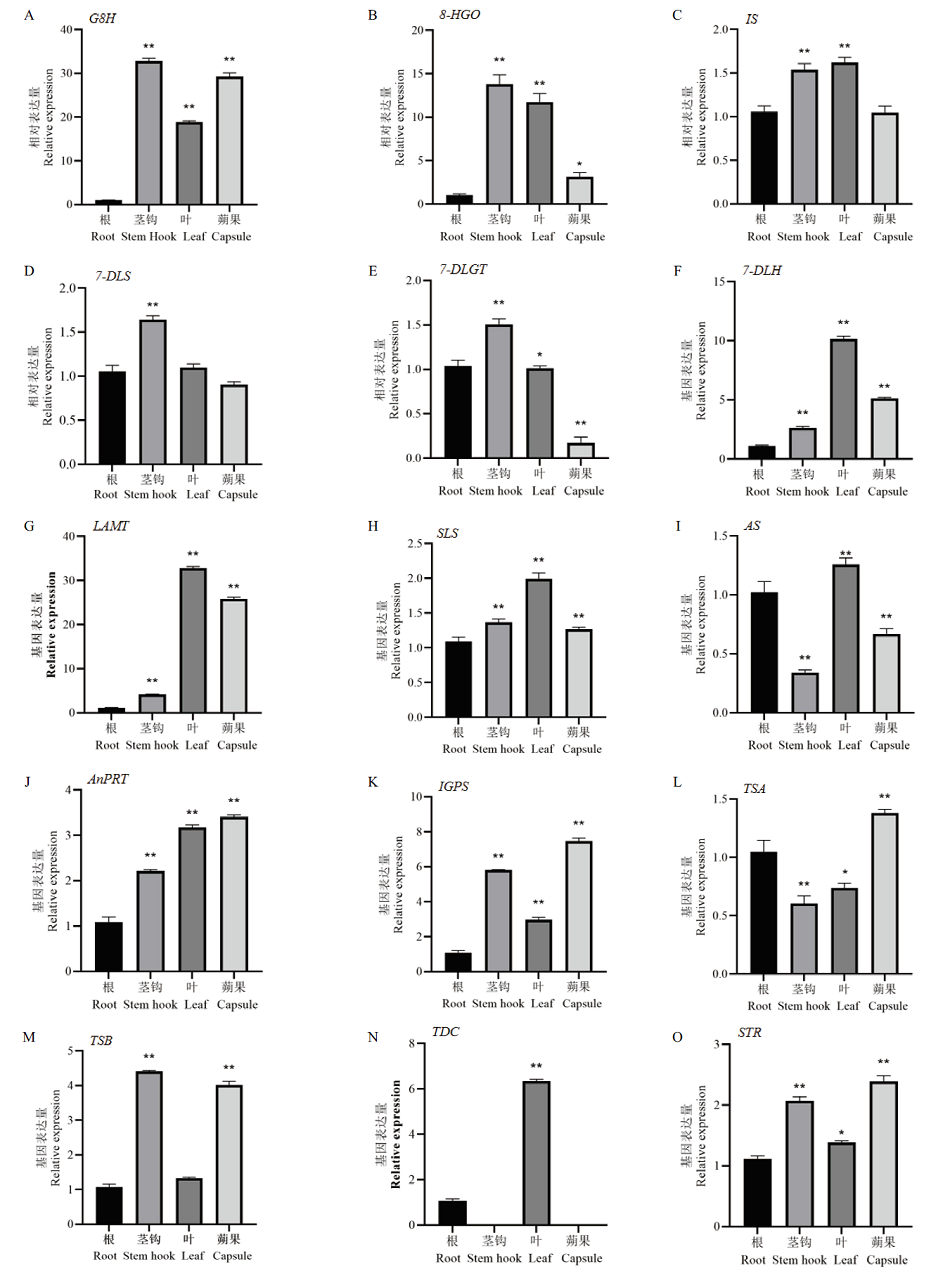
图10 以SAM为管家基因时钩藤生物碱生物合成上游途径中关键酶基因表达模式分析
Fig. 10 Expression analysis of upstream pathway genes involved in TIAs biosynthesis using SAM as house-keeping gene
| [1] | 国家药典委员会. 中国药典[M]. 北京: 中国医药科技出版社, 2020, 268. |
| Chinese Pharmacopoeia Commission. Pharmacopoeia of the People’s Republic of China.[M]. Beijing: China Medical Science Press, 2020: 268. | |
| [2] | 高晓宇, 丁茹, 王道平, 等. 钩藤化学成分及药理作用研究进展[J]. 天津医科大学学报, 2017, 23(4): 380-382. |
| Gao XY, Ding R, Wang DP, et al. Advances in chemical constituents and pharmacological action of Uncaria rhynchophylla.[J]. J Tianjin Med Univ, 2017, 23(4): 380-382. | |
| [3] | 顾雄华, 刘芊, 贺小芳. 天麻钩藤饮合小柴胡汤治疗围绝经期高血压合并焦虑状态临床观察[J]. 北京中医药, 2012, 31(4): 305-308. |
| Gu XH, Liu Q, He XF. Clinical observation of tianma gouteng decoction combined with xiaochaihu decoction in the treatment of perimenopausal hypertension combined with anxiety[J]. Beijing J Tradit Chin Med, 2012, 31(4): 305-308. | |
| [4] | 周吉银, 周世文, 贺燕. 异钩藤碱药理作用的最新研究进展[J]. 中成药, 2013, 35(3): 596-599. |
| Zhou JY, Zhou SW, He Y. The latest research progress of pharmacological action of Isorhynchophylline[J]. Chin Tradit Pat Med, 2013, 35(3): 596-599. | |
| [5] |
Li HL, Wang XB, Liu Y, et al. Hepatoprotection and hepatotoxicity of Heshouwu, a Chinese medicinal herb: context of the paradoxical effect[J]. Food Chem Toxicol, 2017, 108(Pt B): 407-418.
doi: S0278-6915(16)30264-2 pmid: 27484243 |
| [6] | 柳威, 邓林华, 赵英强. 钩藤提取物及钩藤碱的药理研究进展[J]. 中药新药与临床药理, 2021, 32(6): 899-904. |
| Liu W, Deng LH, Zhao YQ. Research progress on pharmacological effects of Uncaria extract and rhynchophylline[J]. Tradit Chin Drug Res Clin Pharmacol, 2021, 32(6): 899-904. | |
| [7] |
Pan HQ, Yang WZ, Zhang YB, et al. An integrated strategy for the systematic characterization and discovery of new indole alkaloids from Uncaria rhynchophylla by UHPLC/DAD/LTQ-Orbitrap-MS[J]. Anal Bioanal Chem, 2015, 407(20): 6057-6070.
doi: 10.1007/s00216-015-8777-0 URL |
| [8] |
Wei SG, Luo ZL, Cui SR, et al. Molecular identification and targeted quantitative analysis of medicinal materials from Uncaria species by DNA barcoding and LC-MS/MS[J]. Molecules, 2019, 24(1): 175.
doi: 10.3390/molecules24010175 URL |
| [9] |
Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays[J]. J Mol Endocrinol, 2000, 25(2): 169-193.
doi: 10.1677/jme.0.0250169 pmid: 11013345 |
| [10] |
Mahoney DJ, Carey K, Fu MH, et al. Real-time RT-PCR analysis of housekeeping genes in human skeletal muscle following acute exercise[J]. Physiol Genomics, 2004, 18(2): 226-231.
pmid: 15161965 |
| [11] |
Maroufi A, van Bockstaele E, de Loose M. Validation of reference genes for gene expression analysis in chicory(Cichorium intybus)using quantitative real-time PCR[J]. BMC Mol Biol, 2010, 11: 15.
doi: 10.1186/1471-2199-11-15 pmid: 20156357 |
| [12] | 易小哲, 邬兰, 向丽, 等. 艾Artemisia argyi实时荧光定量PCR内参基因筛选[J]. 中国中药杂志, 2022, 47(3): 659-667. |
| Yi XZ, Wu L, Xiang L, et al. Screening of reference genes for quantitative real-time PCR in Artemisia argyi[J]. China J Chin Mater Med, 2022, 47(3): 659-667. | |
| [13] | 涂冬萍, 莫长明, 马小军, 等. 罗汉果实时荧光定量PCR内参基因的选择[J]. 中国中药杂志, 2015, 40(2): 204-209. |
| Tu DP, Mo CM, Ma XJ, et al. Selection of reference genes of Siraitia grosvenorii by real-time PCR[J]. China J Chin Mater Med, 2015, 40(2): 204-209. | |
| [14] | 杨婷, 薛珍珍, 李娜, 等. 铁十字秋海棠斑叶发育过程内参基因筛选及验证[J]. 园艺学报, 2021, 48(11): 2251-2261. |
| Yang T, Xue ZZ, Li N, et al. Reference genes selection and validation in Begonia masoniana leaves of different developmental stages[J]. Acta Hortic Sin, 2021, 48(11): 2251-2261. | |
| [15] | 郭茜茜. 钩藤碱和异钩藤碱生物合成路径的研究[D]. 哈尔滨: 东北农业大学, 2014. |
| Guo QQ. Research on rhynchophylline and isorhynchophylline biosynthetic pathway[D]. Harbin: Northeast Agricultural University, 2014. | |
| [16] |
Contin A, van der Heijden R, Lefeber AW, et al. The iridoid glucoside secologanin is derived from the novel triose phosphate/pyruvate pathway in a Catharanthus roseus cell culture[J]. FEBS Lett, 1998, 434(3): 413-416.
doi: 10.1016/s0014-5793(98)01022-9 pmid: 9742965 |
| [17] |
Yamazaki Y, Kitajima M, Arita M, et al. Biosynthesis of camptothecin. in silico and in vivo tracer study from[1-13C]glucose[J]. Plant Physiol, 2004, 134(1): 161-170.
pmid: 14657405 |
| [18] |
Collu G, Unver N, Peltenburg-Looman AM, et al. Geraniol 10-hydroxylase, a cytochrome P450 enzyme involved in terpenoid indole alkaloid biosynthesis[J]. FEBS Lett, 2001, 508(2): 215-220.
doi: 10.1016/s0014-5793(01)03045-9 pmid: 11718718 |
| [19] |
Collu G, Alonso Garcia A, van der Heijden R, et al. Activity of the cytochrome P450 enzyme geraniol 10-hydroxylase and alkaloid production in plant cell cultures[J]. Plant Sci, 2002, 162(1): 165-172.
doi: 10.1016/S0168-9452(01)00554-4 URL |
| [20] |
Yamamoto H, Katano N, Ooi A, et al. Secologanin synthase which catalyzes the oxidative cleavage of loganin into secologanin is a cytochrome P450[J]. Phytochemistry, 2000, 53(1): 7-12.
pmid: 10656401 |
| [21] |
Irmler S, Schroder G, St-Pierre B, et al. Indole alkaloid biosynthesis in Catharanthus roseus: new enzyme activities and identification of cytochrome P450 CYP72A1 as secologanin synthase[J]. Plant J, 2000, 24(6): 797-804.
doi: 10.1046/j.1365-313x.2000.00922.x pmid: 11135113 |
| [22] |
Stöckigt J, Barleben L, Panjikar S, et al. 3D-Structure and function of strictosidine synthase—the key enzyme of monoterpenoid indole alkaloid biosynthesis[J]. Plant Physiol Biochem, 2008, 46(3): 340-355.
doi: 10.1016/j.plaphy.2007.12.011 URL |
| [23] | Vandesompele J, de Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes[J]. Genome Biol, 2002, 3(7): RESEARCH0034. |
| [24] |
Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets[J]. Cancer Res, 2004, 64(15): 5245-5250.
doi: 10.1158/0008-5472.CAN-04-0496 pmid: 15289330 |
| [25] |
Pfaffl MW, Tichopad A, Prgomet C, et al. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations[J]. Biotechnol Lett, 2004, 26(6): 509-515.
doi: 10.1023/B:BILE.0000019559.84305.47 URL |
| [26] |
Xie F, Xiao P, Chen D, et al. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs[J]. Plant Mol Biol, 2012, 80(1): 75-84.
doi: 10.1007/s11103-012-9885-2 URL |
| [27] | 姚李祥, 潘春柳, 余丽莹, 等. 草果种子休眠解除过程中RT-qPCR内参基因筛选[J]. 中国中药杂志, 2021, 46(15): 3832-3837. |
| Yao LX, Pan CL, Yu LY, et al. Selection of RT-qPCR reference genes for Amomum tsaoko seeds during dormancy release[J]. China J Chin Mater Med, 2021, 46(15): 3832-3837. | |
| [28] | 吴建阳, 何冰, 杜玉洁, 等. 利用geNorm、NormFinder和BestKeeper软件进行内参基因稳定性分析的方法[J]. 现代农业科技, 2017(5): 278-281. |
| Wu JY, He B, Du YJ, et al. Analysis method of systematically evaluating stability of reference genes using geNorm, NormFinder and BestKeeper[J]. Mod Agric Sci Technol, 2017(5): 278-281. | |
| [29] |
Tang Q, Ma XJ, Mo CM, et al. An efficient approach to finding Siraitia grosvenorii triterpene biosynthetic genes by RNA-seq and digital gene expression analysis[J]. BMC Genomics, 2011, 12: 343.
doi: 10.1186/1471-2164-12-343 pmid: 21729270 |
| [30] |
Dai LH, Liu C, Zhu YM, et al. Functional characterization of cucurbitadienol synthase and triterpene glycosyltransferase involved in biosynthesis of mogrosides from Siraitia grosvenorii[J]. Plant Cell Physiol, 2015, 56(6): 1172-1182.
doi: 10.1093/pcp/pcv043 URL |
| [31] |
Zhang JS, Dai LH, Yang JG, et al. Oxidation of cucurbitadienol catalyzed by CYP87D18 in the biosynthesis of mogrosides from Siraitia grosvenorii[J]. Plant Cell Physiol, 2016, 57(5): 1000-1007.
doi: 10.1093/pcp/pcw038 URL |
| [32] | 陈凌艳, 谢德金, 荣俊冬, 等. 花叶唐竹4种叶色表型RT-qPCR内参基因筛选[J]. 分子植物育种, 2019, 17(14): 4592-4599. |
| Chen LY, Xie DJ, Rong JD, et al. Screening of RT-qPCR internal reference genes for four leaf color phenotypes of Sinobambusa tootsik f. luteoloalbostriata[J]. Mol Plant Breed, 2019, 17(14): 4592-4599. | |
| [33] | 代红军, 秦晨亮, 徐伟荣. 赤霞珠葡萄发育后期RT-PCR内参基因的筛选和验证[J]. 江苏农业学报, 2016, 32(3): 668-673. |
| Dai HJ, Qin CL, Xu WR. Screening and validation of reference genes for real-time fluorescence quantitative PCR during grape berry development of Cabernet Sauvignon[J]. Jiangsu J Agric Sci, 2016, 32(3): 668-673. | |
| [34] | 杨坤, 黄超, 卢山, 等. 铜胁迫下紫鸭跖草根组织实时定量PCR内参基因的选择[J]. 植物生理学报, 2021, 57(1): 195-204. |
| Yang K, Huang C, Lu S, et al. Reference gene selection for quantitative real-time PCR in purple setcreasea(Setcreasea purpurea)root tissue under copper stress[J]. Plant Physiol J, 2021, 57(1): 195-204. | |
| [35] |
Geu-Flores F, Sherden NH, Courdavault V, et al. An alternative route to cyclic terpenes by reductive cyclization in iridoid biosynthesis[J]. Nature, 2012, 492(7427): 138-142.
doi: 10.1038/nature11692 URL |
| [36] | 吴昕怡, 刘小莉. 环烯醚萜类成分生物合成途径及关键酶基因研究进展[J]. 中国民族民间医药, 2017, 26(8): 44-48. |
| Wu XY, Liu XL. Progress of biosynthetic pathway and the key enzyme genes of iridoids[J]. Chin J Ethnomedicine Ethnopharmacy, 2017, 26(8): 44-48. | |
| [37] |
Noé W, Mollenschott C, Berlin J. Tryptophan decarboxylase from Catharanthus roseus cell suspension cultures: purification, molecular and kinetic data of the homogenous protein[J]. Plant Mol Biol, 1984, 3(5): 281-288.
doi: 10.1007/BF00017782 pmid: 24310513 |
| [1] | 林红妍, 郭晓蕊, 刘迪, 李慧, 陆海. 转录组分析转录因子AtbHLH68调控细胞壁发育的分子机制[J]. 生物技术通报, 2023, 39(9): 105-116. |
| [2] | 娄慧, 朱金成, 杨洋, 张薇. 抗、感品种棉花根系分泌物对尖孢镰刀菌生长及基因表达的影响[J]. 生物技术通报, 2023, 39(9): 156-167. |
| [3] | 杨志晓, 侯骞, 刘国权, 卢志刚, 曹毅, 芶剑渝, 王轶, 林英超. 不同抗性烟草品系Rubisco及其活化酶对赤星病胁迫的响应[J]. 生物技术通报, 2023, 39(9): 202-212. |
| [4] | 陈中元, 王玉红, 代为俊, 张艳敏, 叶倩, 刘旭平, 谭文松, 赵亮. 柠檬酸铁铵对悬浮HEK293细胞转染的影响机制探究[J]. 生物技术通报, 2023, 39(9): 311-318. |
| [5] | 王佳蕊, 孙培媛, 柯瑾, 冉彬, 李洪有. 苦荞糖基转移酶基因FtUGT143的克隆及表达分析[J]. 生物技术通报, 2023, 39(8): 204-212. |
| [6] | 李博, 刘合霞, 陈宇玲, 周兴文, 朱宇林. 金花茶CnbHLH79转录因子的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 241-250. |
| [7] | 付钰, 贾瑞瑞, 何荷, 王良桂, 杨秀莲. 两种砧木楸树嫁接苗生长差异及转录组比较分析[J]. 生物技术通报, 2023, 39(8): 251-261. |
| [8] | 王帅, 冯宇梅, 白苗, 杜维俊, 岳爱琴. 大豆GmHMGR基因响应外源激素及非生物胁迫功能研究[J]. 生物技术通报, 2023, 39(7): 131-142. |
| [9] | 孙明慧, 吴琼, 刘丹丹, 焦小雨, 王文杰. 茶树CsTMFs的克隆与表达分析[J]. 生物技术通报, 2023, 39(7): 151-159. |
| [10] | 余慧, 王静, 梁昕昕, 辛亚平, 周军, 赵会君. 宁夏枸杞铁镉响应基因的筛选及其功能验证[J]. 生物技术通报, 2023, 39(7): 195-205. |
| [11] | 梅欢, 李玥, 刘可蒙, 刘吉华. 小檗碱桥酶高效原核表达及生物合成l-SLR的研究[J]. 生物技术通报, 2023, 39(7): 277-287. |
| [12] | 赵雪婷, 高利燕, 王俊刚, 沈庆庆, 张树珍, 李富生. 甘蔗AP2/ERF转录因子基因ShERF3的克隆、表达及其编码蛋白的定位[J]. 生物技术通报, 2023, 39(6): 208-216. |
| [13] | 张路阳, 韩文龙, 徐晓雯, 姚健, 李芳芳, 田效园, 张智强. 烟草TCP基因家族的鉴定及表达分析[J]. 生物技术通报, 2023, 39(6): 248-258. |
| [14] | 杨洋, 朱金成, 娄慧, 韩泽刚, 张薇. 海岛棉与枯萎病菌的互作转录组分析[J]. 生物技术通报, 2023, 39(6): 259-273. |
| [15] | 李帜奇, 袁月, 苗荣庆, 庞秋颖, 张爱琴. 盐胁迫盐芥和拟南芥褪黑素含量及合成相关基因表达模式分析[J]. 生物技术通报, 2023, 39(5): 142-151. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||