








生物技术通报 ›› 2024, Vol. 40 ›› Issue (2): 233-244.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0874
收稿日期:2023-09-11
出版日期:2024-02-26
发布日期:2024-03-13
通讯作者:
伍国强,男,博士,教授,博士生导师,研究方向:植物逆境生理与基因工程;E-mail: gqwu@lut.edu.cn作者简介:李昊,男,硕士研究生,研究方向:植物逆境生理与基因工程;E-mail: 1044624636@qq.com
基金资助:
LI Hao( ), WU Guo-qiang(
), WU Guo-qiang( ), WEI Ming, HAN Yue-xin
), WEI Ming, HAN Yue-xin
Received:2023-09-11
Published:2024-02-26
Online:2024-03-13
摘要:
【目的】甜菜碱醛脱氢酶(betaine aldehyde dehydrogenase, BADH)是参与甘氨酸甜菜碱(glycine betaine, GB)合成的关键酶之一,在植物响应逆境胁迫过程中发挥重要作用。【方法】为探究甜菜(Beta vulgaris)BvBADH 基因家族成员生物学功能及基因表达模式,本研究采用生物信息学方法,分析了蛋白质的理化性质、系统进化、基因结构、染色体定位、保守基序、蛋白结构、启动子顺式作用调控元件,并通过RT-qPCR对其在盐胁迫下的表达模式进行分析。【结果】共鉴定出9个BvBADH 基因家族成员,含有9-15个外显子,8-14个内含子,平均氨基酸个数为516,平均分子量为55.84 kD,等电点为5.24-6.98。系统进化分析发现,高等植物BADH可分为3个簇,分别为簇I、II和III,其中簇III成员进一步分为簇IIIa和IIIb两个亚簇。BvBADH基因家族在3个簇中均有分布,分别含有3、1和5个成员。甜菜BvBADH基因家族主要含有胁迫响应元件、激素响应元件和生长发育响应元件,每个成员的顺式作用调控元件数量为13-21个。进一步对 BvBADH在盐处理下甜菜叶片中的表达模式分析发现,100和150 mmol/L NaCl不同程度地诱导BvBADH基因家族成员的表达;但相比之下,它们在100 mmol/L NaCl处理下的表达量高于150 mmol/L NaCl。【结论】BvBADH基因家族在甜菜响应盐胁迫过程中扮演着重要角色,为我国北方地区农作物抗逆性遗传改良提供优异基因资源。
李昊, 伍国强, 魏明, 韩悦欣. 甜菜BvBADH基因家族全基因组鉴定及其高盐胁迫下的表达分析[J]. 生物技术通报, 2024, 40(2): 233-244.
LI Hao, WU Guo-qiang, WEI Ming, HAN Yue-xin. Genome-wide Identification of the BvBADH Gene Family in Sugar Beet(Beta vulgaris)and Their Expression Analysis Under High Salt Stress[J]. Biotechnology Bulletin, 2024, 40(2): 233-244.
| 基因Gene | 正向引物序列Forward primer sequence(5'-3') | 反向引物序列Reverse primer sequence(5'-3') |
|---|---|---|
| BvBADH1 | GCAGTGCAACATCCAGACTC | CCCCCTCACTCTTTGCTGTT |
| BvBADH2 | GTCCTGTTGTCAGCAAGGGA | GCATGGACGTGGAGACATCA |
| BvBADH3 | CCAAATTGGGGGAAGTTTGTGT | TGGATCACCAACAACCCAAGA |
| BvBADH4 | ACGGGGAATTTGTGGATGCT | TCATCACCCTTCCCCTTTCA |
| BvBADH5 | ATTGAGCATGGCAAGCGAGA | TGCCATTCGCTCGCTCTATC |
| BvBADH6 | GCCGATTTAGCACCACTTCT | GCAGAAGTGGTGCTAAATCGG |
| BvBADH7 | GCATCTGATTATGGGCTGGC | CCACTCATTTTGTACCCGCC |
| BvBADH8 | TCGTCGAGCCAATAACACCC | CCACCAAATGGAATGCCTGC |
| BvBADH9 | GGCCAGTTCAGAGGTGAGGA | GAACTTGGACCCAAGCCTTTT |
| BvACTIN | ACTGGTATTGTGCTTGACTC | ATGAGATAATCAGTGAGATC |
表1 本研究所用RT-qPCR引物序列
Table 1 Sequences of primers used for RT-qPCR in this study
| 基因Gene | 正向引物序列Forward primer sequence(5'-3') | 反向引物序列Reverse primer sequence(5'-3') |
|---|---|---|
| BvBADH1 | GCAGTGCAACATCCAGACTC | CCCCCTCACTCTTTGCTGTT |
| BvBADH2 | GTCCTGTTGTCAGCAAGGGA | GCATGGACGTGGAGACATCA |
| BvBADH3 | CCAAATTGGGGGAAGTTTGTGT | TGGATCACCAACAACCCAAGA |
| BvBADH4 | ACGGGGAATTTGTGGATGCT | TCATCACCCTTCCCCTTTCA |
| BvBADH5 | ATTGAGCATGGCAAGCGAGA | TGCCATTCGCTCGCTCTATC |
| BvBADH6 | GCCGATTTAGCACCACTTCT | GCAGAAGTGGTGCTAAATCGG |
| BvBADH7 | GCATCTGATTATGGGCTGGC | CCACTCATTTTGTACCCGCC |
| BvBADH8 | TCGTCGAGCCAATAACACCC | CCACCAAATGGAATGCCTGC |
| BvBADH9 | GGCCAGTTCAGAGGTGAGGA | GAACTTGGACCCAAGCCTTTT |
| BvACTIN | ACTGGTATTGTGCTTGACTC | ATGAGATAATCAGTGAGATC |
| 基因名称 Gene name | 基因登录号 Gene ID | CDS /bp | 相对分子量 Mw/kD | 亲水性平均值 GRAVY | 等电点 pI | 脂肪指数 Aliphatic index | 氨基酸数目 Protein length/aa | 不稳定指数 Instability index |
|---|---|---|---|---|---|---|---|---|
| BvBADH1 | Bv5_ 116250_aodi | 1 500 | 54.72 | -0.077 | 5.45 | 88.78 | 500 | 32.48 |
| BvBADH2 | Bv5_ 116230_ntjn | 1 512 | 54.78 | -0.062 | 5.37 | 88.51 | 503 | 31.62 |
| BvBADH3 | Bv8_ 195870_rfyd | 1 509 | 54.69 | -0.081 | 5.24 | 87.45 | 502 | 28.42 |
| BvBADH4 | Bv8_ 195860_yiyp | 1 506 | 54.25 | -0.079 | 5.60 | 89.00 | 501 | 22.07 |
| BvBADH5 | Bv8_ 195850_giqy | 1 509 | 54.34 | -0.039 | 6.01 | 88.05 | 502 | 25.97 |
| BvBADH6 | Bv5_ 116750_zqhy | 1 605 | 58.11 | -0.111 | 6.65 | 85.90 | 534 | 31.08 |
| BvBADH7 | Bv6_ 149040_dgrz | 1 605 | 57.84 | -0.076 | 6.53 | 89.55 | 534 | 32.99 |
| BvBADH8 | Bv6_ 149060_fmfu | 1 608 | 58.71 | -0.109 | 6.54 | 85.74 | 535 | 32.04 |
| BvBADH9 | Bv2_047750_ktxe | 1 602 | 57.11 | 0.066 | 6.98 | 95.72 | 533 | 38.12 |
表2 甜菜BvBADH基因家族成员鉴定
Table 2 Identification of BvBADH gene family members in sugar beet
| 基因名称 Gene name | 基因登录号 Gene ID | CDS /bp | 相对分子量 Mw/kD | 亲水性平均值 GRAVY | 等电点 pI | 脂肪指数 Aliphatic index | 氨基酸数目 Protein length/aa | 不稳定指数 Instability index |
|---|---|---|---|---|---|---|---|---|
| BvBADH1 | Bv5_ 116250_aodi | 1 500 | 54.72 | -0.077 | 5.45 | 88.78 | 500 | 32.48 |
| BvBADH2 | Bv5_ 116230_ntjn | 1 512 | 54.78 | -0.062 | 5.37 | 88.51 | 503 | 31.62 |
| BvBADH3 | Bv8_ 195870_rfyd | 1 509 | 54.69 | -0.081 | 5.24 | 87.45 | 502 | 28.42 |
| BvBADH4 | Bv8_ 195860_yiyp | 1 506 | 54.25 | -0.079 | 5.60 | 89.00 | 501 | 22.07 |
| BvBADH5 | Bv8_ 195850_giqy | 1 509 | 54.34 | -0.039 | 6.01 | 88.05 | 502 | 25.97 |
| BvBADH6 | Bv5_ 116750_zqhy | 1 605 | 58.11 | -0.111 | 6.65 | 85.90 | 534 | 31.08 |
| BvBADH7 | Bv6_ 149040_dgrz | 1 605 | 57.84 | -0.076 | 6.53 | 89.55 | 534 | 32.99 |
| BvBADH8 | Bv6_ 149060_fmfu | 1 608 | 58.71 | -0.109 | 6.54 | 85.74 | 535 | 32.04 |
| BvBADH9 | Bv2_047750_ktxe | 1 602 | 57.11 | 0.066 | 6.98 | 95.72 | 533 | 38.12 |
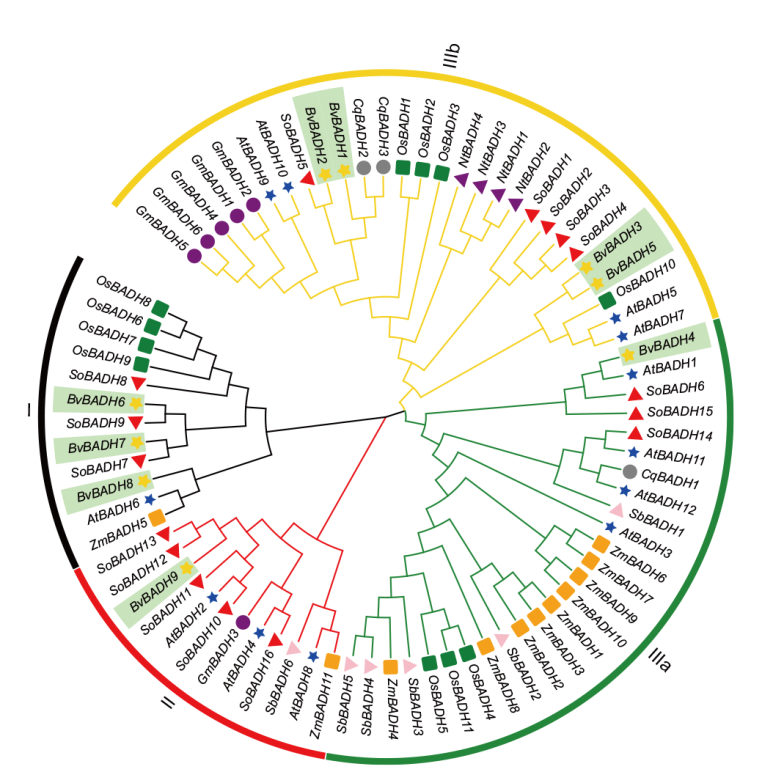
图2 植物BADH基因家族系统发育树 BADHs分为3个簇,簇I(黑色)、簇II(红色)和簇III ,其中簇III 又分为簇III a(绿色)和III b(金色);深绿色标记为甜菜BvBADH家族成员
Fig. 2 Phylogenetic tree of BADH gene family in plants BADHs is divided into three clusters. Cluster I(black), II(red)and III. Cluster III is further divided into Cluster III a(green)and III b(gold). The dark green marks in the figure represent the BvBADH family members

图3 甜菜BvBADH基因家族氨基酸序列多重比对 蓝色方框为保守十肽序列
Fig. 3 Amino acid sequence multiple alignment of the BvBADH gene family in sugar beet The blue box is a conserved decapeptide sequence
| 顺式作用元件Cis-acting element | 功能Function | 序列Sequence | BvBADH | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ||||
| ABRE | Abscisic acid-responsive element | ACGTG | 3 | 2 | 5 | 3 | 3 | 2 | 0 | 4 | 3 | |
| TCA-element | Involved in salicylic acid responsiveness | CCATCTTTTT | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 0 | 1 | |
| TGA-element | Auxin-responsive element | AACGAC | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | |
| ARE | Anaerobic-responsive element | AAACCA | 4 | 3 | 1 | 2 | 4 | 4 | 5 | 2 | 6 | |
| LTR | Low-temperature responsiveness | CCGAAA | 1 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 2 | |
| Box 4 | Involved in light responsiveness | ATTAAT | 3 | 3 | 2 | 1 | 0 | 1 | 0 | 3 | 1 | |
| GT1-motif | Involved in light responsiveness | GGTTAAT | 2 | 0 | 4 | 3 | 4 | 0 | 2 | 2 | 1 | |
| TCT-motif | Involved in light responsiveness | TCTTAC | 6 | 0 | 2 | 1 | 1 | 1 | 1 | 2 | 0 | |
| AE-box | Modul for light response | AGAAACTT | 0 | 1 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | |
| O2-site | Zein metabolism regulation | GATGACATGA | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | |
| G-box | Involved in light responsiveness | TACGTG | 1 | 2 | 7 | 0 | 4 | 4 | 3 | 0 | 0 | |
表3 BvBADHs基因启动子顺式作用调控元件分析
Table 3 Analysis of cis-acting regulatory elements in the promoter of BvBADHs genes
| 顺式作用元件Cis-acting element | 功能Function | 序列Sequence | BvBADH | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ||||
| ABRE | Abscisic acid-responsive element | ACGTG | 3 | 2 | 5 | 3 | 3 | 2 | 0 | 4 | 3 | |
| TCA-element | Involved in salicylic acid responsiveness | CCATCTTTTT | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 0 | 1 | |
| TGA-element | Auxin-responsive element | AACGAC | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | |
| ARE | Anaerobic-responsive element | AAACCA | 4 | 3 | 1 | 2 | 4 | 4 | 5 | 2 | 6 | |
| LTR | Low-temperature responsiveness | CCGAAA | 1 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 2 | |
| Box 4 | Involved in light responsiveness | ATTAAT | 3 | 3 | 2 | 1 | 0 | 1 | 0 | 3 | 1 | |
| GT1-motif | Involved in light responsiveness | GGTTAAT | 2 | 0 | 4 | 3 | 4 | 0 | 2 | 2 | 1 | |
| TCT-motif | Involved in light responsiveness | TCTTAC | 6 | 0 | 2 | 1 | 1 | 1 | 1 | 2 | 0 | |
| AE-box | Modul for light response | AGAAACTT | 0 | 1 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | |
| O2-site | Zein metabolism regulation | GATGACATGA | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | |
| G-box | Involved in light responsiveness | TACGTG | 1 | 2 | 7 | 0 | 4 | 4 | 3 | 0 | 0 | |
| 蛋白名称 Protein name | α螺旋 α-helix | 伸展主链 Extended strand | β转角 β-turn | 无规卷曲 Random coil |
|---|---|---|---|---|
| BvBADH1 | 43.40 | 16.40 | 7.80 | 32.40 |
| BvBADH2 | 42.94 | 15.90 | 7.55 | 33.60 |
| BvBADH3 | 40.84 | 17.53 | 8.17 | 33.47 |
| BvBADH4 | 40.32 | 17.56 | 7.98 | 34.13 |
| BvBADH5 | 39.44 | 17.73 | 8.37 | 34.46 |
| BvBADH6 | 43.07 | 15.54 | 7.49 | 33.90 |
| BvBADH7 | 41.76 | 15.54 | 7.87 | 34.83 |
| BvBADH8 | 42.80 | 16.07 | 7.85 | 33.27 |
| BvBADH9 | 48.22 | 15.01 | 8.07 | 28.71 |
表4 BvBADHs蛋白二级结构组成
Table 4 Secondary structure composition of BvBADHs proteins %
| 蛋白名称 Protein name | α螺旋 α-helix | 伸展主链 Extended strand | β转角 β-turn | 无规卷曲 Random coil |
|---|---|---|---|---|
| BvBADH1 | 43.40 | 16.40 | 7.80 | 32.40 |
| BvBADH2 | 42.94 | 15.90 | 7.55 | 33.60 |
| BvBADH3 | 40.84 | 17.53 | 8.17 | 33.47 |
| BvBADH4 | 40.32 | 17.56 | 7.98 | 34.13 |
| BvBADH5 | 39.44 | 17.73 | 8.37 | 34.46 |
| BvBADH6 | 43.07 | 15.54 | 7.49 | 33.90 |
| BvBADH7 | 41.76 | 15.54 | 7.87 | 34.83 |
| BvBADH8 | 42.80 | 16.07 | 7.85 | 33.27 |
| BvBADH9 | 48.22 | 15.01 | 8.07 | 28.71 |

图6 BvBADHs蛋白质二级结构 蓝色为α螺旋;红色为伸展主链;绿色为β转角;紫色为无规卷曲
Fig. 6 Secondary structure of BvBADHs protein Blue is the α-helix; red is the extended strand; green is the β-turn; purple is the random coil
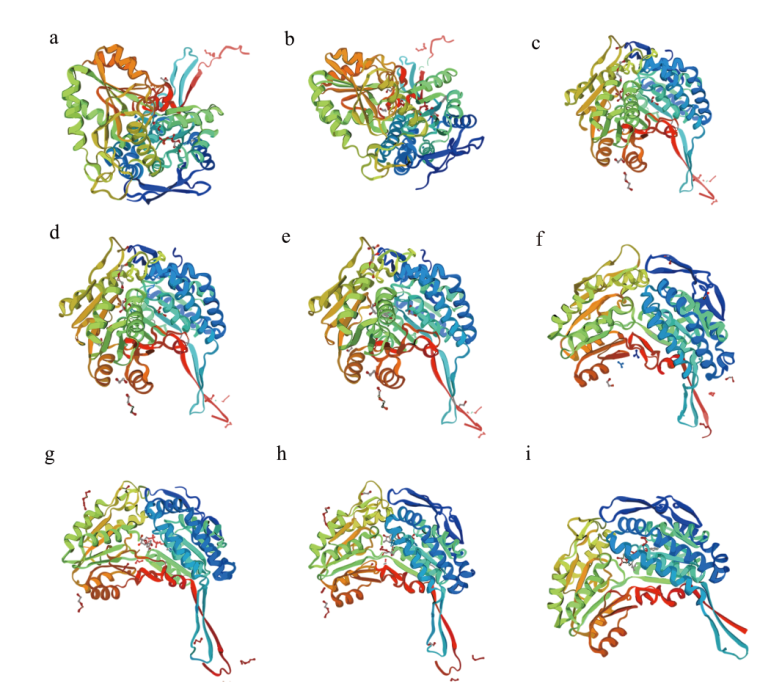
图7 甜菜BvBADH蛋白质三维结构 a-i:BvBADH1-BvBADH9三维结构示意图
Fig. 7 Three-dimensional structure of BvBADH proteins in sugar beet a-i: Schematic diagram of the three-dimensional structure of BvBADH1-BvBADH9

图8 不同浓度 NaCl 处理下甜菜叶片BvBADHs相对表达水平 竖线表示标准误差(SE),n=3。误差线上小写字母表示在P<0.05水平差异显著
Fig. 8 Relative expressions of BvBADHs in sugar beet leaves under various concentrations of NaCl The vertical bar represents the standard error(SE), n=3. The lowercase letter at the top of each bar indicates a significant deviation at the P<0.05 level
| [1] |
Zhang HM, Zhu JH, Gong ZZ, et al. Abiotic stress responses in plants[J]. Nat Rev Genet, 2022, 23(2): 104-119.
doi: 10.1038/s41576-021-00413-0 |
| [2] |
Singh A. Soil salinization management for sustainable development: a review[J]. J Environ Manage, 2021, 277: 111383.
doi: 10.1016/j.jenvman.2020.111383 URL |
| [3] |
Singh A. Alternative management options for irrigation-induced salinization and waterlogging under different climatic conditions[J]. Ecol Indic, 2018, 90: 184-192.
doi: 10.1016/j.ecolind.2018.03.014 URL |
| [4] |
Hazen SP, Wu YJ, Kreps JA. Gene expression profiling of plant responses to abiotic stress[J]. Funct Integr Genomics, 2003, 3(3): 105-111.
doi: 10.1007/s10142-003-0088-4 URL |
| [5] |
Kumar S, Dhingra A, Daniell H. Plastid-expressed betaine aldehyde dehydrogenase gene in carrot cultured cells, roots, and leaves confers enhanced salt tolerance[J]. Plant Physiol, 2004, 136(1): 2843-2854.
doi: 10.1104/pp.104.045187 pmid: 15347789 |
| [6] |
Zhou SF, Chen XY, Zhang XG, et al. Improved salt tolerance in tobacco plants by co-transformation of a betaine synthesis gene BADH and a vacuolar Na+/H+ antiporter gene SeNHX1[J]. Biotechnol Lett, 2008, 30(2): 369-376.
doi: 10.1007/s10529-007-9548-6 URL |
| [7] |
Chen THH, Murata N. Enhancement of tolerance of abiotic stress by metabolic engineering of betaines and other compatible solutes[J]. Curr Opin Plant Biol, 2002, 5(3): 250-257.
doi: 10.1016/s1369-5266(02)00255-8 pmid: 11960744 |
| [8] |
Shahbazi M, Tohidfar M, Aliniaeifard S, et al. Transgenic tobacco co-expressing flavodoxin and betaine aldehyde dehydrogenase confers cadmium tolerance through boosting antioxidant capacity[J]. Protoplasma, 2022, 259(4): 965-979.
doi: 10.1007/s00709-021-01714-1 |
| [9] | 刘振林, 戴思兰. 植物甜菜碱醛脱氢酶基因研究进展[J]. 西北农林科技大学学报: 自然科学版, 2004, 32(3): 104-112. |
| Liu ZL, Dai SL. Studies on betaine aldehyde dehydrogenase(BADH)gene in plants[J]. J Northwest Sci Tech Univ Agric For, 2004, 32(3): 104-112. | |
| [10] |
Abid M, Gu SC, Zhang YJ, et al. Comparative transcriptome and metabolome analysis reveal key regulatory defense networks and genes involved in enhanced salt tolerance of Actinidia(kiwifruit)[J]. Hortic Res, 2022, 9: uhac189.
doi: 10.1093/hr/uhac189 URL |
| [11] |
Mitsuya S, Kuwahara J, Ozaki K, et al. Isolation and characterization of a novel peroxisomal choline monooxygenase in barley[J]. Planta, 2011, 234(6): 1215-1226.
doi: 10.1007/s00425-011-1478-9 pmid: 21769646 |
| [12] |
Yang XH, Liang Z, Wen XG, et al. Genetic engineering of the biosynthesis of glycinebetaine leads to increased tolerance of photosynthesis to salt stress in transgenic tobacco plants[J]. Plant Mol Biol, 2008, 66(1/2): 73-86.
doi: 10.1007/s11103-007-9253-9 URL |
| [13] |
Omari AF. Metabolic engineering of osmoprotectants to elucidate the mechanism(s)of salt stress tolerance in crop plants[J]. Planta, 2021, 253(1): 24.
doi: 10.1007/s00425-020-03550-8 |
| [14] |
Nakamura T, Nomura M, Mori H, et al. An isozyme of betaine aldehyde dehydrogenase in barley[J]. Plant Cell Physiol, 2001, 42(10): 1088-1092.
pmid: 11673624 |
| [15] |
Weretilnyk EA, Hanson AD. Betaine aldehyde dehydrogenase from spinach leaves: purification, in vitro translation of the mRNA, and regulation by salinity[J]. Arch Biochem Biophys, 1989, 271(1): 56-63.
pmid: 2712575 |
| [16] |
Ishitani M, Nakamura T, Han SY, et al. Expression of the betaine aldehyde dehydrogenase gene in barley in response to osmotic stress and abscisic acid[J]. Plant Mol Biol, 1995, 27(2): 307-315.
doi: 10.1007/BF00020185 pmid: 7888620 |
| [17] |
Wood AJ, Saneoka H, Rhodes D, et al. Betaine aldehyde dehydrogenase in sorghum(molecular cloning and expression of two related genes)[J]. Plant Physiol, 1996, 110(4): 1301-1308.
doi: 10.1104/pp.110.4.1301 pmid: 8934627 |
| [18] |
Nakamura T, Yokota S, Muramoto Y, et al. Expression of a betaine aldehyde dehydrogenase gene in rice, a glycinebetaine nonaccumulator, and possible localization of its protein in peroxisomes[J]. Plant J, 1997, 11(5): 1115-1120.
doi: 10.1046/j.1365-313x.1997.11051115.x pmid: 9193078 |
| [19] |
Legaria J, Rajsbaum R, Muñoz-Clares RA, et al. Molecular characterization of two genes encoding betaine aldehyde dehydrogenase from amaranth. Expression in leaves under short-term exposure to osmotic stress or abscisic acid[J]. Gene, 1998, 218(1/2): 69-76.
doi: 10.1016/S0378-1119(98)00381-3 URL |
| [20] |
Hibino T, Meng YL, Kawamitsu Y, et al. Molecular cloning and functional characterization of two kinds of betaine-aldehyde dehydrogenase in betaine-accumulating mangrove Avicennia marina(Forsk.)Vierh[J]. Plant Mol Biol, 2001, 45(3): 353-363.
pmid: 11292080 |
| [21] | Yin XJ, Zhao YX, Luo D, et al. Isolating the promoter of a stress-induced gene encoding betaine aldehyde dehydrogenase from the halophyte Atriplex centralasiatica Iljin[J]. Biochim Biophys Acta, 2002, 1577(3): 452-456. |
| [22] |
Li QL, Gao XR, Yu XH, et al. Molecular cloning and characterization of betaine aldehyde dehydrogenase gene from Suaeda liaotun-gensis and its use in improved tolerance to salinity in transgenic tobacco[J]. Biotechnol Lett, 2003, 25(17): 1431-1436.
doi: 10.1023/A:1025003628446 URL |
| [23] |
Rezaei Qusheh Bolagh F, Solouki A, Tohidfar M, et al. Agrobacte-rium-mediated transformation of Persian walnut using BADH gene for salt and drought tolerance[J]. J Hortic Sci Biotechnol, 2021, 96(2): 162-171.
doi: 10.1080/14620316.2020.1812446 URL |
| [24] |
Di H, Tian Y, Zu HY, et al. Enhanced salinity tolerance in transgenic maize plants expressing a BADH gene from Atriplex micran-tha[J]. Euphytica, 2015, 206(3): 775-783.
doi: 10.1007/s10681-015-1515-z URL |
| [25] |
Ali A, Ali Q, Iqbal MS, et al. Salt tolerance of potato genetically engineered with the Atriplex canescens BADH gene[J]. Biol Plant, 2020, 64: 271-279.
doi: 10.32615/bp.2019.080 URL |
| [26] |
Liu ZH, Zhang HM, Li GL, et al. Enhancement of salt tolerance in alfalfa transformed with the gene encoding for betaine aldehyde dehydrogenase[J]. Euphytica, 2011, 178(3): 363-372.
doi: 10.1007/s10681-010-0316-7 URL |
| [27] |
Chaum S, Supaibulwatana K, Kirdmanee C. Glycinebetaine accumulation, physiological characterizations and growth efficiency in salt-tolerant and salt-sensitive lines of indica rice(Oryza sativa L. ssp. indica)in response to salt stress[J]. J Agron Crop Sci, 2007, 193(3): 157-166.
doi: 10.1111/jac.2007.193.issue-3 URL |
| [28] |
Li PF, Cai J, Luo X, et al. Transformation of wheat Triticum aes-tivum with the HvBADH1 transgene from hulless barley improves salinity-stress tolerance[J]. Acta Physiol Plant, 2019, 41(9): 155.
doi: 10.1007/s11738-019-2940-8 |
| [29] | Wang FW, Wang ML, Guo C, et al. Cloning and characterization of a novel betaine aldehyde dehydrogenase gene from Suaeda cornic-ulata[J]. Genet Mol Res, 2016, 15(2): 15027848. |
| [30] |
Sun YL, Liu X, Fu L, et al. Overexpression of TaBADH increases salt tolerance in Arabidopsis[J]. Can J Plant Sci, 2019, 99: 546-555.
doi: 10.1139/cjps-2018-0190 URL |
| [31] |
Wakeel A, Asif AR, Pitann B, et al. Proteome analysis of sugar beet(Beta vulgaris L.)elucidates constitutive adaptation during the first phase of salt stress[J]. J Plant Physiol, 2011, 168(6): 519-526.
doi: 10.1016/j.jplph.2010.08.016 URL |
| [32] |
Liu H, Wang QQ, Yu MM, et al. Transgenic salt-tolerant sugar beet(Beta vulgaris L.)constitutively expressing an Arabidopsis thalia-na vacuolar Na/H antiporter gene, AtNHX3, accumulates more soluble sugar but less salt in storage roots[J]. Plant Cell Environ, 2008, 31(9): 1325-1334.
doi: 10.1111/pce.2008.31.issue-9 URL |
| [33] | Gasteiger E, Hoogland C, Gattiker A, et al. Protein identification and analysis tools on the ExPASy server[M]//The Proteomics Protocols Handbook. Totowa, NJ: Humana Press, 2005: 571-607. |
| [34] |
Kumar S, Stecher G, Li M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms[J]. Mol Biol Evol, 2018, 35(6): 1547-1549.
doi: 10.1093/molbev/msy096 pmid: 29722887 |
| [35] |
Hu B, Jin JP, Guo AY, et al. GSDS 2.0: an upgraded gene feature visualization server[J]. Bioinformatics, 2015, 31(8): 1296-1297.
doi: 10.1093/bioinformatics/btu817 pmid: 25504850 |
| [36] |
Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers[J]. Proc Int Conf Intell Syst Mol Biol, 1994, 2: 28-36.
pmid: 7584402 |
| [37] |
Lescot M, Déhais P, Thijs G, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences[J]. Nucleic Acids Res, 2002, 30(1): 325-327.
doi: 10.1093/nar/30.1.325 pmid: 11752327 |
| [38] |
Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0[J]. Bioinformatics, 2007, 23(21): 2947-2948.
doi: 10.1093/bioinformatics/btm404 pmid: 17846036 |
| [39] |
Pan SM, Moreau RA, Yu C, et al. Betaine accumulation and betaine-aldehyde dehydrogenase in spinach leaves[J]. Plant Physiol, 1981, 67(6): 1105-1108.
doi: 10.1104/pp.67.6.1105 pmid: 16661818 |
| [40] |
Weigel P, Weretilnyk EA, Hanson AD. Betaine aldehyde oxidation by spinach chloroplasts[J]. Plant Physiol, 1986, 82(3): 753-759.
doi: 10.1104/pp.82.3.753 pmid: 16665106 |
| [41] |
Min MH, Maung TZ, Cao Y, et al. Haplotype analysis of BADH1 by next-generation sequencing reveals association with salt tolerance in rice during domestication[J]. Int J Mol Sci, 2021, 22(14): 7578.
doi: 10.3390/ijms22147578 URL |
| [42] |
Wang SB, Liang HP, Wang HL, et al. The chromosome-scale genomes of Dipterocarpus turbinatus and Hopea hainanensis(Dipterocarpaceae)provide insights into fragrant oleoresin biosynthesis and hardwood formation[J]. Plant Biotechnol J, 2022, 20(3): 538-553.
doi: 10.1111/pbi.v20.3 URL |
| [43] | 武映宏, 黄东益, 王永, 等. 椰子ALDH基因家族的鉴定及生物信息学分析[J]. 热带生物学报, 2022, 13(6): 541-549. |
| Wu YH, Huang DY, Wang Y, et al. Identification and bioinformatics analysis of ALDH gene family in coconut[J]. J Trop Biol, 2022, 13(6): 541-549. | |
| [44] |
Basanagouda G, Ramesh S, Siddu CB, et al. A non-synonymous SNP in homolog of BADH2 gene is associated with fresh pod fragrance in dolichos bean(Lablab purpureus var. lignosus(Prain)Kumari)[J]. Genet Resour Crop Evol, 2023, 70(2): 373-380.
doi: 10.1007/s10722-022-01535-y |
| [45] |
Luo ML, Sun HJ, Ge WY, et al. Effect of glycine betaine treatment on aroma production of ‘Nanguo’ pears after long-term cold storage-possible involvement of ethylene synthesis and signal transduction pathways[J]. Food Bioprocess Technol, 2022, 15(6): 1327-1342.
doi: 10.1007/s11947-022-02813-4 |
| [46] |
Saensuk C, Ruangnam S, Pitaloka MK, et al. A SNP of betaine aldehyde dehydrogenase(BADH)enhances an aroma(2-acetyl-1-pyrroline)in sponge gourd(Luffa cylindrica)and ridge gourd(Luffa acutangula)[J]. Sci Rep, 2022, 12(1): 3718.
doi: 10.1038/s41598-022-07478-9 |
| [47] |
Moghaieb REA, Saneoka H, Fujita K. Effect of salinity on osmotic adjustment, glycinebetaine accumulation and the betaine aldehyde dehydrogenase gene expression in two halophytic plants, Salicornia europaea and Suaeda maritima[J]. Plant Sci, 2004, 166(5): 1345-1349.
doi: 10.1016/j.plantsci.2004.01.016 URL |
| [48] | Senthilkumar M, Amaresan N, Sankaranarayanan A. Estimation of betaine aldehyde dehydrogenase(BADH)activity[M]// Plant-Microbe Interactions. New York: Humana, 2021: 101-102. |
| [49] | 何晓兰, 侯喜林, 吴纪中, 等. 三角叶滨藜甜菜碱醛脱氢酶(BADH)基因的克隆及序列分析[J]. 南京农业大学学报, 2004, 27(1): 15-19. |
| He XL, Hou XL, Wu JZ, et al. Molecular cloning and sequence analysis of betaine-aldehyde dehydrogenase(BADH)in Atriplex triangularis[J]. J Nanjing Agric Univ, 2004, 27(1): 15-19. | |
| [50] | 陈翠萍. 藜麦BADH基因家族鉴定及生物信息学分析[J]. 分子植物育种, 2023, 21(19): 6276-6284. |
| Chen CP. Identification and bioinformatics analysis of BADH gene family in quinoa[J]. Mol Plant Breed, 2023, 21(19): 6276-6284. | |
| [51] | 刘振林, 曹华雯, 夏新莉, 等. 甘菊BADH基因cDNA的克隆及在盐胁迫下的表达[J]. 武汉植物学研究, 2009, 27(1): 1-7. |
| Liu ZL, Cao HW, Xia XL, et al. Cloning and expression analysis of betaine aldehyde dehydrogenase gene from Dendranthema lavan-dulifolium on salinity[J]. J Wuhan Bot Res, 2009, 27(1): 1-7. | |
| [52] | 唐露, 杨振, 白蓓蓓, 等. 香蕉BADH基因序列及表达特性分析[J]. 分子植物育种, 2019, 17(3): 719-728. |
| Tang L, Yang Z, Bai BB, et al. Sequence analysis and expression characteristics of BADH gene in banana[J]. Mol Plant Breed, 2019, 17(3): 719-728. | |
| [53] |
Ding X, Li JH, Pan Y, et al. Genome-wide identification and expression analysis of the UGlcAE gene family in tomato[J]. Int J Mol Sci, 2018, 19(6): 1583.
doi: 10.3390/ijms19061583 URL |
| [54] |
Liu Z, Ge XY, Yang ZR, et al. Genome-wide identification and characterization of SnRK2 gene family in cotton(Gossypium hirsu-tum L.)[J]. BMC Genet, 2017, 18(1): 54.
doi: 10.1186/s12863-017-0517-3 URL |
| [55] |
Zhang Y, Yin H, Li D, et al. Functional analysis of BADH gene promoter from Suaeda liaotungensis K[J]. Plant Cell Rep, 2008, 27(3): 585-592.
doi: 10.1007/s00299-007-0459-8 pmid: 17924116 |
| [56] |
Li CY, Zhang TP, Feng P, et al. Genetic engineering of glycinebetaine synthesis enhances cadmium tolerance in BADH-transgenic tobacco plants via reducing cadmium uptake and alleviating cadmium stress damage[J]. Environ Exp Bot, 2021, 191: 104602.
doi: 10.1016/j.envexpbot.2021.104602 URL |
| [57] |
Shahzad A, Qian MC, Sun BY, et al. Genome-wide association study identifies novel loci and candidate genes for drought stress tolerance in rapeseed[J]. Oil Crop Sci, 2021, 6(1): 12-22.
doi: 10.1016/j.ocsci.2021.01.002 URL |
| [58] |
Zhang L, Tian JJ, Ye LZ, et al. Transcriptomic analysis reveals the mechanism of low/high temperature resistance in an outstanding diet alga Nannochloropsis oceanica[J]. Aquaculture Reports, 2022, 27: 101365.
doi: 10.1016/j.aqrep.2022.101365 URL |
| [59] |
Ming RH, Zhang Y, Wang Y, et al. The JA-responsive MYC2-BADH-like transcriptional regulatory module in Poncirus trifoliata contributes to cold tolerance by modulation of glycine betaine biosynthesis[J]. New Phytol, 2021, 229(5): 2730-2750.
doi: 10.1111/nph.v229.5 URL |
| [60] |
Feng CG, Liu RY, Xu WJ, et al. The genome of a new anemone species(Actiniaria: Hormathiidae)provides insights into deep-sea adaptation[J]. Deep Sea Res Part I Oceanogr Res Pap, 2021, 170:103492.
doi: 10.1016/j.dsr.2021.103492 URL |
| [61] |
Jacques F, Zhao YJ, Kopečná M, et al. Roles for ALDH10 enzymes in γ-butyrobetaine synthesis, seed development, germination, and salt tolerance in Arabidopsis[J]. J Exp Bot, 2020, 71(22): 7088-7102.
doi: 10.1093/jxb/eraa394 URL |
| [1] | 徐扬, 张瑞英, 戴良香, 张冠初, 丁红, 张智猛. 盐胁迫下氮素对花生种子萌发和种子际细菌菌群结构的调控[J]. 生物技术通报, 2024, 40(2): 253-265. |
| [2] | 王雨晴, 马子奇, 侯嘉欣, 宗钰琪, 郝晗睿, 刘国元, 魏辉, 连博琳, 陈艳红, 张健. 盐胁迫下植物根系分泌物的成分分析与生态功能研究进展[J]. 生物技术通报, 2024, 40(1): 12-23. |
| [3] | 焦进兰, 王文文, 介欣芮, 王华忠, 岳洁瑜. 外源钙缓解小麦幼苗盐胁迫的作用机制[J]. 生物技术通报, 2024, 40(1): 207-221. |
| [4] | 王帅, 冯宇梅, 白苗, 杜维俊, 岳爱琴. 大豆GmHMGR基因响应外源激素及非生物胁迫功能研究[J]. 生物技术通报, 2023, 39(7): 131-142. |
| [5] | 魏茜雅, 秦中维, 梁腊梅, 林欣琪, 李映志. 褪黑素种子引发处理提高朝天椒耐盐性的作用机制[J]. 生物技术通报, 2023, 39(7): 160-172. |
| [6] | 张和臣, 袁欣, 高杰, 王校晨, 王慧娟, 李艳敏, 王利民, 符真珠, 李保印. 植物花瓣呈色机理及花色分子育种[J]. 生物技术通报, 2023, 39(5): 23-31. |
| [7] | 王海龙, 李雨倩, 王勃, 邢国芳, 张杰伟. 谷子SiMAPK3基因的克隆和表达特性分析[J]. 生物技术通报, 2023, 39(3): 123-132. |
| [8] | 杜清洁, 周璐瑶, 杨思震, 张嘉欣, 陈春林, 李娟起, 李猛, 赵士文, 肖怀娟, 王吉庆. 过表达CaCP1提高转基因烟草对盐胁迫的敏感性[J]. 生物技术通报, 2023, 39(2): 172-182. |
| [9] | 叶红, 王玉昆. 植物PRR免疫受体功能研究进展[J]. 生物技术通报, 2023, 39(12): 1-15. |
| [10] | 汪明滔, 刘建伟, 赵春钊. 植物调控盐胁迫下细胞壁完整性的分子机制[J]. 生物技术通报, 2023, 39(11): 18-27. |
| [11] | 张玉娟, 黎冬华, 宫慧慧, 崔新晓, 高春华, 张秀荣, 游均, 赵军胜. 芝麻NAC转录因子基因SiNAC77的克隆及耐盐功能分析[J]. 生物技术通报, 2023, 39(11): 308-317. |
| [12] | 徐扬, 丁红, 张冠初, 郭庆, 张智猛, 戴良香. 盐胁迫下花生种子萌发期代谢组学分析[J]. 生物技术通报, 2023, 39(1): 199-213. |
| [13] | 陈光, 李佳, 杜瑞英, 王旭. 水稻盐敏感突变体ss2的鉴定与基因功能分析[J]. 生物技术通报, 2022, 38(9): 158-166. |
| [14] | 张斌, 杨昕霞. 水稻响应盐胁迫关键转录因子的鉴定[J]. 生物技术通报, 2022, 38(3): 9-15. |
| [15] | 张业猛, 朱丽丽, 陈志国. 藜麦NHX基因家族鉴定及盐胁迫下表达分析[J]. 生物技术通报, 2022, 38(12): 184-193. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||