








生物技术通报 ›› 2023, Vol. 39 ›› Issue (5): 23-31.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1229
张和臣1( ), 袁欣1, 高杰1, 王校晨1,2, 王慧娟1, 李艳敏1, 王利民1, 符真珠1(
), 袁欣1, 高杰1, 王校晨1,2, 王慧娟1, 李艳敏1, 王利民1, 符真珠1( ), 李保印2(
), 李保印2( )
)
收稿日期:2022-10-08
出版日期:2023-05-26
发布日期:2023-06-08
通讯作者:
符真珠,女,博士,副研究员,研究方向:组培快繁及相关生物学机理;E-mail: pearlgh2005@163.com;作者简介:张和臣,男,博士,副研究员,研究方向:植物花瓣呈色机理及花色育种;E-mail: zhc5128@126.com
基金资助:
ZHANG He-chen1( ), YUAN Xin1, GAO Jie1, WANG Xiao-chen1,2, WANG Hui-juan1, LI Yan-min1, WANG Li-min1, FU Zhen-zhu1(
), YUAN Xin1, GAO Jie1, WANG Xiao-chen1,2, WANG Hui-juan1, LI Yan-min1, WANG Li-min1, FU Zhen-zhu1( ), LI Bao-yin2(
), LI Bao-yin2( )
)
Received:2022-10-08
Published:2023-05-26
Online:2023-06-08
摘要:
植物花瓣呈色的主要化学物质包括类黄酮/花青素苷、类胡萝卜素和甜菜色素。其中类黄酮/花青素苷是分布最广泛的色素,决定大多数植物花瓣的呈色;类胡萝卜素在一些植物黄色至橙红色花瓣中起着作用;而甜菜色素主要存在于石竹目植物,包含甜菜红素和甜菜黄素。目前,关于色素生物合成的分子网络已被解析,主要由一系列结构基因控制;一些与色素合成相关的调控因子在很多植物中被鉴定发现。另外,基于外源基因表达或内源基因编辑的分子育种在一些观赏植物的花色改良中被成功应用。本文系统性总结了植物中3种类型色素合成的分子基础、调控机制及分子育种应用等方面的研究进展;将有助于提高我们对植物色素合成分子调控网络的认识,并以期为今后开展花色分子设计育种提供理论支持。
张和臣, 袁欣, 高杰, 王校晨, 王慧娟, 李艳敏, 王利民, 符真珠, 李保印. 植物花瓣呈色机理及花色分子育种[J]. 生物技术通报, 2023, 39(5): 23-31.
ZHANG He-chen, YUAN Xin, GAO Jie, WANG Xiao-chen, WANG Hui-juan, LI Yan-min, WANG Li-min, FU Zhen-zhu, LI Bao-yin. Mechanism of Flower Petal Coloration and Molecular Breeding[J]. Biotechnology Bulletin, 2023, 39(5): 23-31.
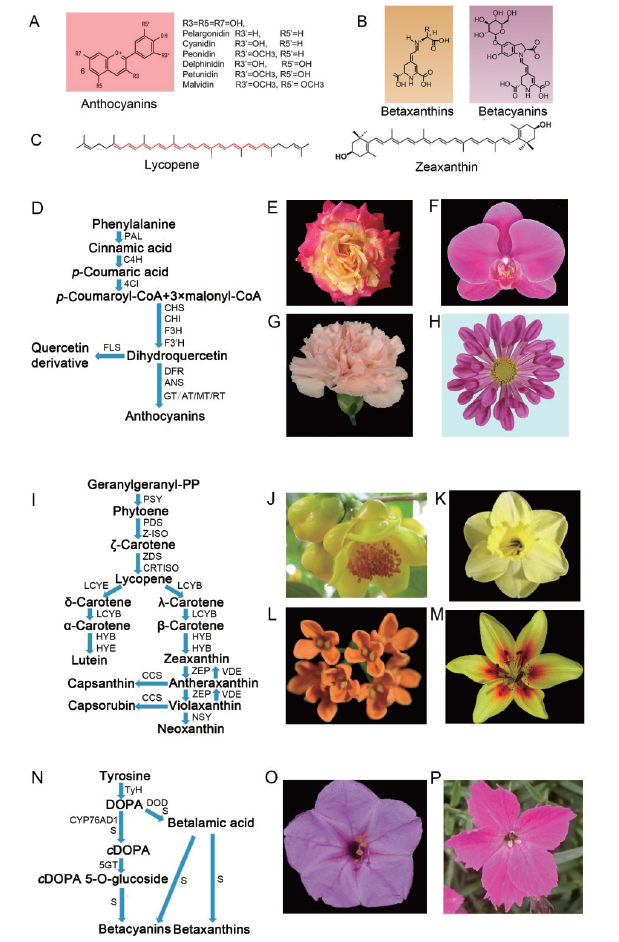
图1 植物不同类型色素化学结构、生物合成途径及代表花卉种类 A-C:分别为花青素苷、甜菜色素及类胡萝卜素化学结构;D:花青素苷生物合成途径;E-H:分别为月季、蝴蝶兰、康乃馨及菊花;I:类胡萝卜素生物合成途径;J-M:分别为金花茶、水仙、桂花及亚洲百合;N:甜菜色素生物合成途径;O-P:分别为紫茉莉及石竹
Fig. 1 Types of plant pigments, biosynthetic pathways and representative flower species A-C: Chemical structure of anthocyanins, beet pigments and carotenoids, respectively. D: Anthocyanins biosynthesis pathway. E-H: The flower petals of Rosa, Phalaenopsis, Dianthus and Chrysanthemum morifolium, respectively. I: Carotenoids biosynthesis pathway. J-M: The flower petals of Camellia aureus, Narcissus, Osmanthus and Lilium asiatica. N: Betalains biosynthesis pathway. O-P: The flower petals of Mirabilis jalapa and Dianthus chinensis, respectively
| [1] |
Rudall PJ. Colourful cones: how did flower colour first evolve?[J]. J Exp Bot, 2020, 71(3): 759-767.
doi: 10.1093/jxb/erz479 pmid: 31714579 |
| [2] |
Vignolini S, Moyroud E, Glover BJ, et al. Analysing photonic structures in plants[J]. J R Soc Interface, 2013, 10(87): 20130394.
doi: 10.1098/rsif.2013.0394 URL |
| [3] |
Tanaka Y, Sasaki N, Ohmiya A. Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids[J]. Plant J, 2008, 54(4): 733-749.
doi: 10.1111/j.1365-313X.2008.03447.x URL |
| [4] |
戴思兰, 洪艳. 基于花青素苷合成和呈色机理的观赏植物花色改良分子育种[J]. 中国农业科学, 2016, 49(3): 529-542.
doi: 10.3864/j.issn.0578-1752.2016.03.011 |
|
Dai SL, Hong Y. Molecular breeding for flower colors modification on ornamental plants based on the mechanism of anthocyanins biosynthesis and coloration[J]. Sci Agric Sin, 2016, 49(3): 529-542.
doi: 10.3864/j.issn.0578-1752.2016.03.011 |
|
| [5] |
Wang LS, Hashimoto F, Shiraishi A, et al. Chemical taxonomy of the Xibei tree peony from China by floral pigmentation[J]. J Plant Res, 2004, 117(1): 47-55.
doi: 10.1007/s10265-003-0130-6 URL |
| [6] |
Bombarely A, Moser M, Amrad A, et al. Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida[J]. Nat Plants, 2016, 2(6): 16074.
doi: 10.1038/nplants.2016.74 |
| [7] |
Zhao YW, Wang CK, Huang XY, et al. Anthocyanin stability and degradation in plants[J]. Plant Signal Behav, 2021, 16(12): 1987767.
doi: 10.1080/15592324.2021.1987767 URL |
| [8] |
Behrens CE, Smith KE, Iancu CV, et al. Transport of anthocyanins and other flavonoids by the Arabidopsis ATP-binding cassette transporter AtABCC2[J]. Sci Rep, 2019, 9(1): 437.
doi: 10.1038/s41598-018-37504-8 pmid: 30679715 |
| [9] |
Sun Y, Li H, Huang JR. Arabidopsis TT19 functions as a carrier to transport anthocyanin from the cytosol to tonoplasts[J]. Mol Plant, 2012, 5(2): 387-400.
doi: 10.1093/mp/ssr110 pmid: 22201047 |
| [10] |
Marinova K, Pourcel L, Weder B, et al. The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H+-antiporter active in proanthocyanidin-accumulating cells of the seed coat[J]. Plant Cell, 2007, 19(6): 2023-2038.
doi: 10.1105/tpc.106.046029 URL |
| [11] |
Zhu CF, Bai C, Sanahuja G, et al. The regulation of carotenoid pigmentation in flowers[J]. Arch Biochem Biophys, 2010, 504(1): 132-141.
doi: 10.1016/j.abb.2010.07.028 pmid: 20688043 |
| [12] |
Sun TH, Yuan H, Cao HB, et al. Carotenoid metabolism in plants: the role of plastids[J]. Mol Plant, 2018, 11(1): 58-74.
doi: S1674-2052(17)30273-3 pmid: 28958604 |
| [13] |
Maoka T. Carotenoids as natural functional pigments[J]. J Nat Med, 2020, 74(1): 1-16.
doi: 10.1007/s11418-019-01364-x |
| [14] |
Yamagishi M, Kishimoto S, Nakayama M. Carotenoid composition and changes in expression of carotenoid biosynthetic genes in tepals of Asiatic hybrid lily[J]. Plant Breed, 2010, 129(1): 100-107.
doi: 10.1111/pbr.2010.129.issue-1 URL |
| [15] |
Moehs CP, Tian L, Osteryoung KW, et al. Analysis of carotenoid biosynthetic gene expression during marigold petal development[J]. Plant Mol Biol, 2001, 45(3): 281-293.
pmid: 11292074 |
| [16] |
Kishimoto S, Ohmiya A. Regulation of carotenoid biosynthesis in petals and leaves of Chrysanthemum(Chrysanthemum morifolium)[J]. Physiol Plant, 2006, 128(3): 436-447.
doi: 10.1111/ppl.2006.128.issue-3 URL |
| [17] |
Yoshioka S, Aida R, Yamamizo C, et al. The carotenoid cleavage dioxygenase 4(CmCCD4a)gene family encodes a key regulator of petal color mutation in Chrysanthemum[J]. Euphytica, 2012, 184(3): 377-387.
doi: 10.1007/s10681-011-0602-z URL |
| [18] |
Lopez AB, van Eck J, Conlin BJ, et al. Effect of the cauliflower Or transgene on carotenoid accumulation and chromoplast formation in transgenic potato tubers[J]. J Exp Bot, 2008, 59(2): 213-223.
doi: 10.1093/jxb/erm299 pmid: 18256051 |
| [19] |
Yamagishi M. How genes paint lily flowers: Regulation of colouration and pigmentation patterning[J]. Sci Hortic, 2013, 163: 27-36.
doi: 10.1016/j.scienta.2013.07.024 URL |
| [20] |
孙叶, 包建忠, 刘春贵, 等. 兰花花色基因工程研究进展[J]. 核农学报, 2015, 29(9): 1701-1710.
doi: 10.11869/j.issn.100-8551.2015.09.1701 |
|
Sun Y, Bao JZ, Liu CG, et al. Progresses on genetic engineering in orchid floral color[J]. J Nucl Agric Sci, 2015, 29(9): 1701-1710.
doi: 10.11869/j.issn.100-8551.2015.09.1701 |
|
| [21] |
Li X, Lu M, Tang DQ, et al. Composition of carotenoids and flavonoids in Narcissus cultivars and their relationship with flower color[J]. PLoS One, 2015, 10(11): e0142074.
doi: 10.1371/journal.pone.0142074 URL |
| [22] |
Zhang C, Wang YG, Fu JX, et al. Transcriptomic analysis and carotenogenic gene expression related to petal coloration in Osmanthus fragrans ‘Yanhong Gui’[J]. Trees, 2016, 30(4): 1207-1223.
doi: 10.1007/s00468-016-1359-8 URL |
| [23] |
Brockington SF, Walker RH, Glover BJ, et al. Complex pigment evolution in the Caryophyllales[J]. New Phytol, 2011, 190(4): 854-864.
doi: 10.1111/j.1469-8137.2011.03687.x pmid: 21714182 |
| [24] |
Timoneda A, Feng T, Sheehan H, et al. The evolution of betalain biosynthesis in Caryophyllales[J]. New Phytol, 2019, 224(1): 71-85.
doi: 10.1111/nph.15980 pmid: 31172524 |
| [25] |
Gandía-Herrero F, García-Carmona F. Biosynthesis of betalains: yellow and violet plant pigments[J]. Trends Plant Sci, 2013, 18(6): 334-343.
doi: 10.1016/j.tplants.2013.01.003 pmid: 23395307 |
| [26] | Mendel G. Gregor mendel's letters to Nägeli[J]. Genetics, 1950, 35:1-29. |
| [27] | Trezzini GF, Zryd JP. Portulaca grandiflora: a model system for the study of the biochemistry and genetics of betalain synthesis[J]. Acta Hortic, 1990(280): 581-585. |
| [28] |
Xu WJ, Dubos C, Lepiniec L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes[J]. Trends Plant Sci, 2015, 20(3): 176-185.
doi: 10.1016/j.tplants.2014.12.001 pmid: 25577424 |
| [29] |
Liu CC, Chi C, Jin LJ, et al. The bZip transcription factor HY5 mediates CRY1a-induced anthocyanin biosynthesis in tomato[J]. Plant Cell Environ, 2018, 41(8): 1762-1775.
doi: 10.1111/pce.v41.8 URL |
| [30] |
An JP, Yao JF, Xu RR, et al. Apple bZIP transcription factor MdbZIP44 regulates abscisic acid-promoted anthocyanin accumulation[J]. Plant Cell Environ, 2018, 41(11): 2678-2692.
doi: 10.1111/pce.v41.11 URL |
| [31] |
Koes R, Verweij W, Quattrocchio F. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways[J]. Trends Plant Sci, 2005, 10(5): 236-242.
doi: 10.1016/j.tplants.2005.03.002 pmid: 15882656 |
| [32] |
Wu Y, Wen J, Xia YP, et al. Evolution and functional diversification of R2R3-MYB transcription factors in plants[J]. Hortic Res, 2022, 9: uhac058.
doi: 10.1093/hr/uhac058 URL |
| [33] |
Albert NW, Lewis DH, Zhang HB, et al. Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning[J]. Plant J, 2011, 65(5): 771-784.
doi: 10.1111/tpj.2011.65.issue-5 URL |
| [34] |
Quattrocchio F, Wing J, van der Woude K, et al. Molecular analysis of the anthocyanin2 gene of Petunia and its role in the evolution of flower color[J]. Plant Cell, 1999, 11(8): 1433-1444.
doi: 10.1105/tpc.11.8.1433 pmid: 10449578 |
| [35] |
Zhang HC, Koes R, Shang HQ, et al. Identification and functional analysis of three new anthocyanin R2R3-MYB genes in Petunia[J]. Plant Direct, 2019, 3(1): e00114.
doi: 10.1002/pld3.114 URL |
| [36] |
Albert NW, Davies KM, Lewis DH, et al. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots[J]. Plant Cell, 2014, 26(3): 962-980.
doi: 10.1105/tpc.113.122069 URL |
| [37] |
Ding BQ, Patterson EL, Holalu SV, et al. Two MYB proteins in a self-organizing activator-inhibitor system produce spotted pigmentation patterns[J]. Curr Biol, 2020, 30(5): 802-814.e8.
doi: S0960-9822(19)31700-2 pmid: 32155414 |
| [38] |
Toledo-Ortiz G, Huq E, Rodríguez-Concepción M. Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors[J]. Proc Natl Acad Sci USA, 2010, 107(25): 11626-11631.
doi: 10.1073/pnas.0914428107 pmid: 20534526 |
| [39] |
Welsch R, Medina J, Giuliano G, et al. Structural and functional characterization of the phytoene synthase promoter from Arabidopsis thaliana[J]. Planta, 2003, 216(3): 523-534.
pmid: 12520345 |
| [40] |
Yin WC, Hu ZL, Cui BL, et al. Suppression of the MADS-box gene SlMBP8 accelerates fruit ripening of tomato(Solanum lycopersicum)[J]. Plant Physiol Biochem, 2017, 118: 235-244.
doi: 10.1016/j.plaphy.2017.06.019 URL |
| [41] |
Liu GY, Ren G, Guirgis A, et al. The MYB305 transcription factor regulates expression of nectarin genes in the ornamental tobacco floral nectary[J]. Plant Cell, 2009, 21(9): 2672-2687.
doi: 10.1105/tpc.108.060079 URL |
| [42] |
Meng YY, Wang ZY, Wang YQ, et al. The MYB activator WHITE PETAL1 associates with MtTT8 and MtWD40-1 to regulate carotenoid-derived flower pigmentation in Medicago truncatula[J]. Plant Cell, 2019, 31(11): 2751-2767.
doi: 10.1105/tpc.19.00480 URL |
| [43] |
Sagawa JM, Stanley LE, LaFountain AM, et al. An R2R3-MYB transcription factor regulates carotenoid pigmentation in Mimulus lewisii flowers[J]. New Phytol, 2016, 209(3): 1049-1057.
doi: 10.1111/nph.2016.209.issue-3 URL |
| [44] |
Wu MB, Xu X, Hu XW, et al. SlMYB72 regulates the metabolism of chlorophylls, carotenoids, and flavonoids in tomato fruit[J]. Plant Physiol, 2020, 183(3): 854-868.
doi: 10.1104/pp.20.00156 pmid: 32414899 |
| [45] |
Polturak G, Aharoni A. “La vie en rose”: biosynthesis, sources, and applications of betalain pigments[J]. Mol Plant, 2018, 11(1): 7-22.
doi: S1674-2052(17)30307-6 pmid: 29081360 |
| [46] |
Hirano H, Sakuta M, Komamine A. Inhibition of betacyanin accumulation by abscisic acid in suspension cultures of Phytolacca americana[J]. Zeitschrift Für Naturforschung C, 1996, 51(11-12): 818-822.
doi: 10.1515/znc-1996-11-1209 URL |
| [47] |
Bianco-Colomas J. Qualitative and quantitative aspects of betalains biosynthesis in Amaranthus caudatus L. var. pendula seedlings[J]. Planta, 1980, 149(2): 176-180.
doi: 10.1007/BF00380880 pmid: 24306250 |
| [48] |
Kochhar VK, Kochhar S, Mohr H. An analysis of the action of light on betalain synthesis in the seedling of Amaranthus caudatus, var. viridis[J]. Planta, 1981, 151(1): 81-87.
doi: 10.1007/BF00384241 pmid: 24301674 |
| [49] |
Chang YC, Chiu YC, Tsao NW, et al. Elucidation of the core betalain biosynthesis pathway in Amaranthus tricolor[J]. Sci Rep, 2021, 11(1): 6086.
doi: 10.1038/s41598-021-85486-x |
| [50] |
Li G, Meng XQ, Zhu MK, et al. Research progress of betalain in response to adverse stresses and evolutionary relationship compared with anthocyanin[J]. Molecules, 2019, 24(17): 3078.
doi: 10.3390/molecules24173078 URL |
| [51] |
Polturak G, Heinig U, Grossman N, et al. Transcriptome and metabolic profiling provides insights into betalain biosynthesis and evolution in Mirabilis jalapa[J]. Mol Plant, 2018, 11(1): 189-204.
doi: S1674-2052(17)30371-4 pmid: 29247705 |
| [52] |
Hatlestad GJ, Akhavan NA, Sunnadeniya RM, et al. The beet Y locus encodes an anthocyanin MYB-like protein that activates the betalain red pigment pathway[J]. Nat Genet, 2015, 47(1): 92-96.
doi: 10.1038/ng.3163 pmid: 25436858 |
| [53] |
Stracke R, Holtgräwe D, Schneider J, et al. Genome-wide identification and characterisation of R2R3-MYB genes in sugar beet(Beta vulgaris)[J]. BMC Plant Biol, 2014, 14: 249.
doi: 10.1186/s12870-014-0249-8 pmid: 25249410 |
| [54] |
Bradley D, Xu P, Mohorianu II, et al. Evolution of flower color pattern through selection on regulatory small RNAs[J]. Science, 2017, 358(6365): 925-928.
doi: 10.1126/science.aao3526 pmid: 29146812 |
| [55] |
Vandenbussche M, Chambrier P, Rodrigues Bento S, et al. Petunia, your next supermodel?[J]. Front Plant Sci, 2016, 7: 72.
doi: 10.3389/fpls.2016.00072 pmid: 26870078 |
| [56] |
Iida S, Hoshino A, Johzuka-Hisatomi Y, et al. Floricultural traits and transposable elements in the Japanese and common morning glories[J]. Ann N Y Acad Sci, 1999, 870: 265-274.
doi: 10.1111/j.1749-6632.1999.tb08887.x URL |
| [57] |
Itoh Y, Higeta D, Suzuki A, et al. Excision of transposable elements from the Chalcone isomerase and dihydroflavonol 4-reductase genes may contribute to the variegation of the yellow-flowered carnation(Dianthus caryophyllus)[J]. Plant Cell Physiol, 2002, 43(5): 578-585.
doi: 10.1093/pcp/pcf065 URL |
| [58] |
Suzuki M, Miyahara T, Tokumoto H, et al. Transposon-mediated mutation of CYP76AD3 affects betalain synthesis and produces variegated flowers in four o'clock(Mirabilis jalapa)[J]. J Plant Physiol, 2014, 171(17): 1586-1590.
doi: 10.1016/j.jplph.2014.07.010 URL |
| [59] |
Gu ZY, Men SQ, Zhu J, et al. Chalcone synthase is ubiquitinated and degraded via interactions with a RING-H2 protein in petals of Paeonia ‘He Xie’[J]. J Exp Bot, 2019, 70(18): 4749-4762.
doi: 10.1093/jxb/erz245 URL |
| [60] |
Napoli C, Lemieux C, Jorgensen R. Introduction of a chimeric Chalcone synthase gene into Petunia results in reversible co-suppression of homologous genes in trans[J]. Plant Cell, 1990, 2(4): 279-289.
doi: 10.2307/3869076 URL |
| [61] |
Tanaka Y, Tsuda S, Kusumi T. Metabolic engineering to modify flower color[J]. Plant Cell Physiol, 1998, 39(11): 1119-1126.
doi: 10.1093/oxfordjournals.pcp.a029312 URL |
| [62] |
Katsumoto Y, Fukuchi-Mizutani M, Fukui Y, et al. Engineering of the rose flavonoid biosynthetic pathway successfully generated blue-hued flowers accumulating delphinidin[J]. Plant Cell Physiol, 2007, 48(11): 1589-1600.
doi: 10.1093/pcp/pcm131 pmid: 17925311 |
| [63] |
Brugliera F, Tao GQ, Tems U, et al. Violet/blue chrysanthemums-metabolic engineering of the anthocyanin biosynthetic pathway results in novel petal colors[J]. Plant Cell Physiol, 2013, 54(10): 1696-1710.
doi: 10.1093/pcp/pct110 pmid: 23926066 |
| [64] |
Nishihara M, Higuchi A, Watanabe A, et al. Application of the CRISPR/Cas9 system for modification of flower color in Torenia fournieri[J]. BMC Plant Biol, 2018, 18(1): 331.
doi: 10.1186/s12870-018-1539-3 pmid: 30518324 |
| [65] |
Yu J, Tu LH, Subburaj S, et al. Simultaneous targeting of duplicated genes in Petunia protoplasts for flower color modification via CRISPR-Cas9 ribonucleoproteins[J]. Plant Cell Rep, 2021, 40(6): 1037-1045.
doi: 10.1007/s00299-020-02593-1 |
| [66] |
Tasaki K, Yoshida M, Nakajima M, et al. Molecular characterization of an anthocyanin-related glutathione S-transferase gene in Japanese Gentian with the CRISPR/Cas9 system[J]. BMC Plant Biol, 2020, 20(1): 370.
doi: 10.1186/s12870-020-02565-3 pmid: 32762648 |
| [67] |
Li YB, Provenzano S, Bliek M, et al. Evolution of tonoplast P-ATPase transporters involved in vacuolar acidification[J]. New Phytol, 2016, 211(3): 1092-1107.
doi: 10.1111/nph.14008 pmid: 27214749 |
| [68] |
Ye X, Al-Babili S, Klöti A, et al. Engineering the provitamin A(beta-carotene)biosynthetic pathway into(carotenoid-free)rice endosperm[J]. Science, 2000, 287(5451): 303-305.
doi: 10.1126/science.287.5451.303 pmid: 10634784 |
| [69] |
Paine JA, Shipton CA, Chaggar S, et al. Improving the nutritional value of Golden Rice through increased pro-vitamin A content[J]. Nat Biotechnol, 2005, 23(4): 482-487.
doi: 10.1038/nbt1082 pmid: 15793573 |
| [70] |
Watkins JL, Pogson BJ. Prospects for carotenoid biofortification targeting retention and catabolism[J]. Trends Plant Sci, 2020, 25(5): 501-512.
doi: S1360-1385(19)30346-2 pmid: 31956035 |
| [71] |
Kishimoto S, Oda-Yamamizo C, Ohmiya A. Comparison of Petunia and Calibrachoa in carotenoid pigmentation of corollas[J]. Breed Sci, 2019, 69(1): 117-126.
doi: 10.1270/jsbbs.18130 URL |
| [72] |
Jeknić Z, Jeknić S, Jevremović S, et al. Alteration of flower color in Iris germanica L. ‘Fire Bride’ through ectopic expression of phytoene synthase gene(crtB)from Pantoea agglomerans[J]. Plant Cell Rep, 2014, 33(8): 1307-1321.
doi: 10.1007/s00299-014-1617-4 pmid: 24801678 |
| [73] |
Polturak G, Grossman N, Vela-Corcia D, et al. Engineered gray mold resistance, antioxidant capacity, and pigmentation in betalain-producing crops and ornamentals[J]. Proc Natl Acad Sci USA, 2017, 114(34): 9062-9067.
doi: 10.1073/pnas.1707176114 pmid: 28760998 |
| [74] |
Gandía-Herrero F, García-Carmona F. Characterization of recombinant beta vulgaris 4, 5-DOPA-extradiol-dioxygenase active in the biosynthesis of betalains[J]. Planta, 2012, 236(1): 91-100.
doi: 10.1007/s00425-012-1593-2 pmid: 22270561 |
| [1] | 吴巧茵, 施友志, 李林林, 彭政, 谭再钰, 刘利平, 张娟, 潘勇. 类胡萝卜素降解菌株的原位筛选及其在雪茄提质增香中的应用[J]. 生物技术通报, 2023, 39(9): 192-201. |
| [2] | 李博, 刘合霞, 陈宇玲, 周兴文, 朱宇林. 金花茶CnbHLH79转录因子的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 241-250. |
| [3] | 王天依, 王荣焕, 王夏青, 张如养, 徐瑞斌, 焦炎炎, 孙轩, 王继东, 宋伟, 赵久然. 玉米矮秆基因与矮秆育种研究[J]. 生物技术通报, 2023, 39(8): 43-51. |
| [4] | 叶云芳, 田清尹, 施婷婷, 王亮, 岳远征, 杨秀莲, 王良桂. 植物中β-紫罗兰酮生物合成及调控研究进展[J]. 生物技术通报, 2023, 39(8): 91-105. |
| [5] | 张蓓, 任福森, 赵洋, 郭志伟, 孙强, 刘贺娟, 甄俊琦, 王童童, 程相杰. 辣椒响应热胁迫机制的研究进展[J]. 生物技术通报, 2023, 39(7): 37-47. |
| [6] | 马芳芳, 刘冠闻, 庞冰, 蒋春美, 师俊玲. 强化细胞外排提高工程菌类黄酮产量的策略[J]. 生物技术通报, 2023, 39(5): 63-76. |
| [7] | 张志霞, 李天培, 曾虹, 朱稀贤, 杨天雄, 马斯楠, 黄磊. 冰冷杆菌PG-2的基因组测序及生物信息学分析[J]. 生物技术通报, 2023, 39(3): 290-300. |
| [8] | 李旺宁, 张豪杰, 李亚男, 梁梦静, 季春丽, 张春辉, 李润植, 崔玉琳, 秦松, 崔红利. 莱茵衣藻蓝光受体植物类型隐花色素CRY突变体的表型鉴定[J]. 生物技术通报, 2023, 39(2): 243-253. |
| [9] | 齐方婷, 黄河. 观赏植物花斑形成调控机制的研究进展[J]. 生物技术通报, 2023, 39(10): 17-28. |
| [10] | 孙威, 张艳, 王聿晗, 徐僡, 徐小蓉, 鞠志刚. 马缨杜鹃Rd3GT1的克隆及对矮牵牛花色形成的影响[J]. 生物技术通报, 2022, 38(9): 198-206. |
| [11] | 周琳, 梁轩铭, 赵磊. 天然类胡萝卜素的生物合成研究进展[J]. 生物技术通报, 2022, 38(7): 119-127. |
| [12] | 刘自然, 甄珍, 陈强, 李玥莹, 王泽, 逄洪波. 植物响应Cd胁迫研究进展[J]. 生物技术通报, 2022, 38(6): 13-26. |
| [13] | 段玥彤, 王鹏年, 张春宝, 林春晶. 植物黄烷酮-3-羟化酶基因研究进展[J]. 生物技术通报, 2022, 38(6): 27-33. |
| [14] | 雷春霞, 李灿辉, 陈永坤, 龚明. 马铃薯块茎形成的生理生化基础和分子机制[J]. 生物技术通报, 2022, 38(4): 44-57. |
| [15] | 田清尹, 岳远征, 申慧敏, 潘多, 杨秀莲, 王良桂. 植物观赏器官中类胡萝卜素代谢调控的研究进展[J]. 生物技术通报, 2022, 38(12): 35-46. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||