








生物技术通报 ›› 2025, Vol. 41 ›› Issue (2): 295-308.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0760
• 研究报告 • 上一篇
向春繁1,2( ), 李勒松1,2, 王娟1,2, 梁艳丽1, 杨生超1,2, 栗孟飞3, 赵艳1,2(
), 李勒松1,2, 王娟1,2, 梁艳丽1, 杨生超1,2, 栗孟飞3, 赵艳1,2( )
)
收稿日期:2024-08-08
出版日期:2025-02-26
发布日期:2025-02-28
通讯作者:
赵艳,女,博士,教授,研究方向 :药用植物学;E-mail: zhaoyankm@126.com作者简介:向春繁,女,硕士研究生,研究方向 :药用植物资源;E-mail: 17869407534@163.com
基金资助:
XIANG Chun-fan1,2( ), LI Le-song1,2, WANG Juan1,2, LIANG Yan-li1, YANG Sheng-chao1,2, LI Meng-fei3, ZHAO Yan1,2(
), LI Le-song1,2, WANG Juan1,2, LIANG Yan-li1, YANG Sheng-chao1,2, LI Meng-fei3, ZHAO Yan1,2( )
)
Received:2024-08-08
Published:2025-02-26
Online:2025-02-28
摘要:
目的 探究当归中木质素合成关键肉桂醇脱氢酶的催化活性及表达特性,为解决当归抽薹后根部木质化问题提供新的研究思路和理论基础。 方法 以非木质化根(UBP)和木质化根(BP)为实验材料,基于转录组数据挖掘候选肉桂醇脱氢酶基因;克隆AsCADs基因的全长CDS,构建pET-28a-AsCADs原核表达载体,转入大肠杆菌(E. coli)BL21(DE3),并诱导重组蛋白表达及体外酶活测定;生物信息学分析具有活性的CAD蛋白序列;利用实时荧光定量PCR对木质化和非木质化根部进行基因表达模式分析。 结果 共鉴定出30个AsCADs基因,其中挖掘并克隆到3个肉桂醇脱氢酶基因AsCAD1、AsCAD4和AsCAD24,均包含2个Zn 2+ 结合位点和1个NAD(H)辅酶结合位点。体外酶活测定发现AsCAD1能催化松柏醛和咖啡醛还原为相应的醇,AsCAD4和AsCAD24能催化对羟基肉桂醛、松柏醛、咖啡醛和芥子醛还原为相应的醇。AsCAD1、AsCAD4和AsCAD24开放阅读框为1 083 bp,编码360个氨基酸,理论等电点(pI)为6.1和5.52,AsCAD1为疏水性蛋白,AsCAD4和AsCAD24为亲水性蛋白,均不含跨膜结构和信号肽,亚细胞定位均定位在细胞质中。RT-qPCR结果显示AsCAD1在非木质化根(UBP)中的表达量高,而AsCAD4和AsCAD24在木质化根(BP)中的表达量高。 结论 成功克隆了AsCAD1、AsCAD4和AsCAD24基因,原核表达后蛋白均具有催化活性,其相对表达水平在木质化与非木质化根中存在差异。
向春繁, 李勒松, 王娟, 梁艳丽, 杨生超, 栗孟飞, 赵艳. 当归肉桂醇脱氢酶AsCAD功能鉴定及表达分析[J]. 生物技术通报, 2025, 41(2): 295-308.
XIANG Chun-fan, LI Le-song, WANG Juan, LIANG Yan-li, YANG Sheng-chao, LI Meng-fei, ZHAO Yan. Functional Identification and Expression Analysis of Cinnamonyl Alcohol Dehydrogenase AsCAD in Angelica sinensis[J]. Biotechnology Bulletin, 2025, 41(2): 295-308.
| 引物名称 Primer name | 引物序列 Primer sequence (5'-3') | 备注 Note |
|---|---|---|
| AsCAD1-F | agcaaatgggtcgcggatccATGGAAAAATCAGCAGAAACCCAAC | 基因克隆 Gene cloning |
| AsCAD1-R | ctcagtggtggtggtggtggtgGGCAGCCTGCAATGTATTTCCA | |
| AsCAD4-F | acagcaaatgggtcgcggatccATGGGCAGCTTGGAAGTG | |
| AsCAD4-R | tcagtggtggtggtggtggtgGGTTTCTTGATCAAGTTTGCTGC | |
| AsCAD24-F | acagcaaatgggtcgcggatccATGGGCAGCTTGGAAGTG | |
| AsCAD24-R | ggtggtggtggtggtgTGTTTCTCGATCAAGTTTGCTACCTG | |
| 环P-pET-28a-F | tggacagcaaatgggtcgcggatcc | 环状载体克隆 Circular vector cloning |
| 环P-pET-28a-R | ggatccgcgacccatttgctgtccac | |
| pET-28a-F-质粒 | atctcgatcccgcgaaattaatac | 原核表达Prokaryotic expression |
| pET-28a-R-质粒 | tcctttcagcaaaaaacccct | |
| q-AsCAD1-F | GGGAGGAAAATAGTAGCCGGTAG | 实时荧光定量PCR RT-qPCR |
| q-AsCAD1-R | CAAGACGTTCCATTGCAGTGTTC | |
| q-AsCAD4-F | GAAAGACGAGGCAATGGATCATC | |
| q-AsCAD4-R | CAACAAGGAGAGGTAAGGTTCGA | |
| q-AsCAD24-F | CACTCGAACCTTACCTCTCCTTG | |
| q-AsCAD24-R | CCCTATGAAGCTCCCTGTTATGG | |
| q-AsEEF1G-F | GTCCCAGCAGCCAAAAAGTC | 内参基因 Reference gene |
| q-AsEEF1G-R | TCTGCCTTGGGCAATTCCTT |
表1 引物序列
Table 1 Primer sequences
| 引物名称 Primer name | 引物序列 Primer sequence (5'-3') | 备注 Note |
|---|---|---|
| AsCAD1-F | agcaaatgggtcgcggatccATGGAAAAATCAGCAGAAACCCAAC | 基因克隆 Gene cloning |
| AsCAD1-R | ctcagtggtggtggtggtggtgGGCAGCCTGCAATGTATTTCCA | |
| AsCAD4-F | acagcaaatgggtcgcggatccATGGGCAGCTTGGAAGTG | |
| AsCAD4-R | tcagtggtggtggtggtggtgGGTTTCTTGATCAAGTTTGCTGC | |
| AsCAD24-F | acagcaaatgggtcgcggatccATGGGCAGCTTGGAAGTG | |
| AsCAD24-R | ggtggtggtggtggtgTGTTTCTCGATCAAGTTTGCTACCTG | |
| 环P-pET-28a-F | tggacagcaaatgggtcgcggatcc | 环状载体克隆 Circular vector cloning |
| 环P-pET-28a-R | ggatccgcgacccatttgctgtccac | |
| pET-28a-F-质粒 | atctcgatcccgcgaaattaatac | 原核表达Prokaryotic expression |
| pET-28a-R-质粒 | tcctttcagcaaaaaacccct | |
| q-AsCAD1-F | GGGAGGAAAATAGTAGCCGGTAG | 实时荧光定量PCR RT-qPCR |
| q-AsCAD1-R | CAAGACGTTCCATTGCAGTGTTC | |
| q-AsCAD4-F | GAAAGACGAGGCAATGGATCATC | |
| q-AsCAD4-R | CAACAAGGAGAGGTAAGGTTCGA | |
| q-AsCAD24-F | CACTCGAACCTTACCTCTCCTTG | |
| q-AsCAD24-R | CCCTATGAAGCTCCCTGTTATGG | |
| q-AsEEF1G-F | GTCCCAGCAGCCAAAAAGTC | 内参基因 Reference gene |
| q-AsEEF1G-R | TCTGCCTTGGGCAATTCCTT |
生物信息学分析 Bioinformatics analysis | 网址 Website links |
|---|---|
| 开放阅读框(ORF) | https://www.ncbi.nlm.nih.gov/orffinder/ORF |
| 蛋白理化性质分析 | https://web.expasy.org/protparam/ |
| 蛋白质二级结构 | https://npsa-prabi.ibcp.fr/cgi-bin/secpredsopma.pl |
| 蛋白质三级结构 | https://swissmodel.expasy.org/interactive |
| 蛋白信号肽预测 | https://services.healthtech.dtu.dk/services/SignalP-6.0/ |
| 蛋白跨膜结构域预测 | https://services.healthtech.dtu.dk/services/TMHMM-2.0/ |
| 蛋白亚细胞定位 | https://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2 |
表2 生物信息学分析网站链接
Table 2 Website links for bioinformatics analysis
生物信息学分析 Bioinformatics analysis | 网址 Website links |
|---|---|
| 开放阅读框(ORF) | https://www.ncbi.nlm.nih.gov/orffinder/ORF |
| 蛋白理化性质分析 | https://web.expasy.org/protparam/ |
| 蛋白质二级结构 | https://npsa-prabi.ibcp.fr/cgi-bin/secpredsopma.pl |
| 蛋白质三级结构 | https://swissmodel.expasy.org/interactive |
| 蛋白信号肽预测 | https://services.healthtech.dtu.dk/services/SignalP-6.0/ |
| 蛋白跨膜结构域预测 | https://services.healthtech.dtu.dk/services/TMHMM-2.0/ |
| 蛋白亚细胞定位 | https://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2 |

图1 当归非木质化根(UBP)和木质化根(BP)番红固绿染色切片
Fig. 1 Safranin-Fast Green staining sections of roots from unbolted plant (UBP) and bolted plant (BP) in Angelica sinensis
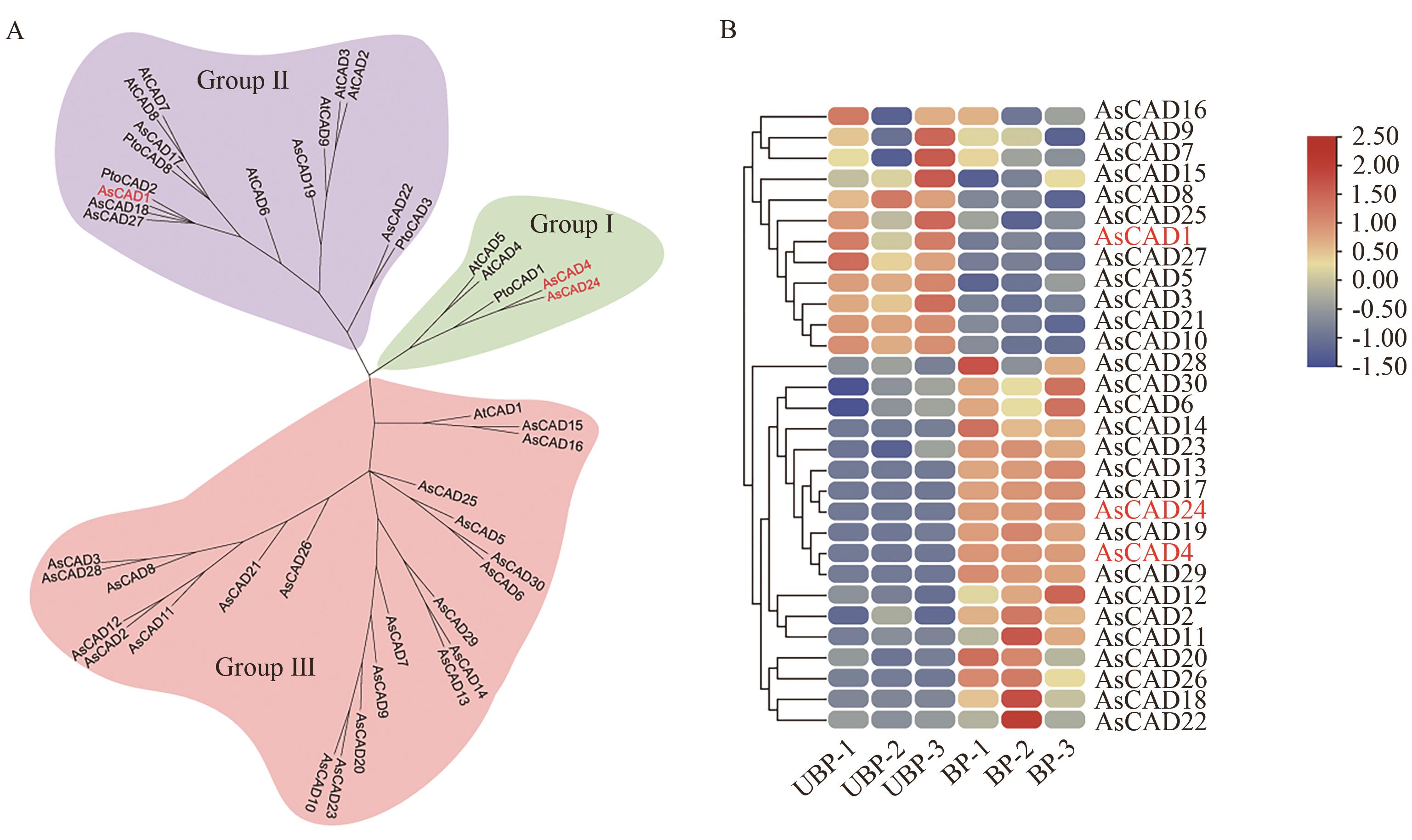
图2 当归肉桂醇脱氢酶基因系统进化树及表达模式分析A:CAD家族进化树分析;B:CAD基因在当归UBP与BP中的表达量热图
Fig. 2 Phylogenetic tree and expression pattern analysis of cinnamyl alcohol dehydrogenase genes in A. sinensisA: Analysis of the CAD family evolutionary tree. B: Heatmap of the expressions of CAD genes in UBP and BP of A. sinensis

图4 AsCAD1、AsCAD4和AsCAD24基因克隆和重组蛋白SDS-PAGE分析M: Marker; A: AsCAD基因全长克隆PCR产物电泳(1,2,3依次为AsCAD1、AsCAD4和AsCAD24基因全长克隆产物); B-D: pET-28a-His-AsCAD重组蛋白SDS-PAGE分析(B: pET-28a-His-AsCAD1复性上清; C: pET-28a-His-AsCAD4复性上清;D: pET-28a-His-AsCAD24复性上清)
Fig. 4 Cloning of the AsCAD1, AsCAD4 and AsCAD24 genes and SDS-PAGE analysis of the recombinant proteinsM: Marker. A: Electrophoresis of full-length cloning PCR products of AsCAD genes (1, 2, and 3 are the full-length cloning products of AsCAD1, AsCAD4, and AsCAD24 genes, respectively). B-D: SDS-PAGE analysis of pET-28a-His-AsCAD recombinant proteins (B: Renatured supernatant of pET-28a-His-AsCAD1. C: Renatured supernatant of pET-28a-His-AsCAD4. D: Renatured supernatant of pET-28a-His-AsCAD24)
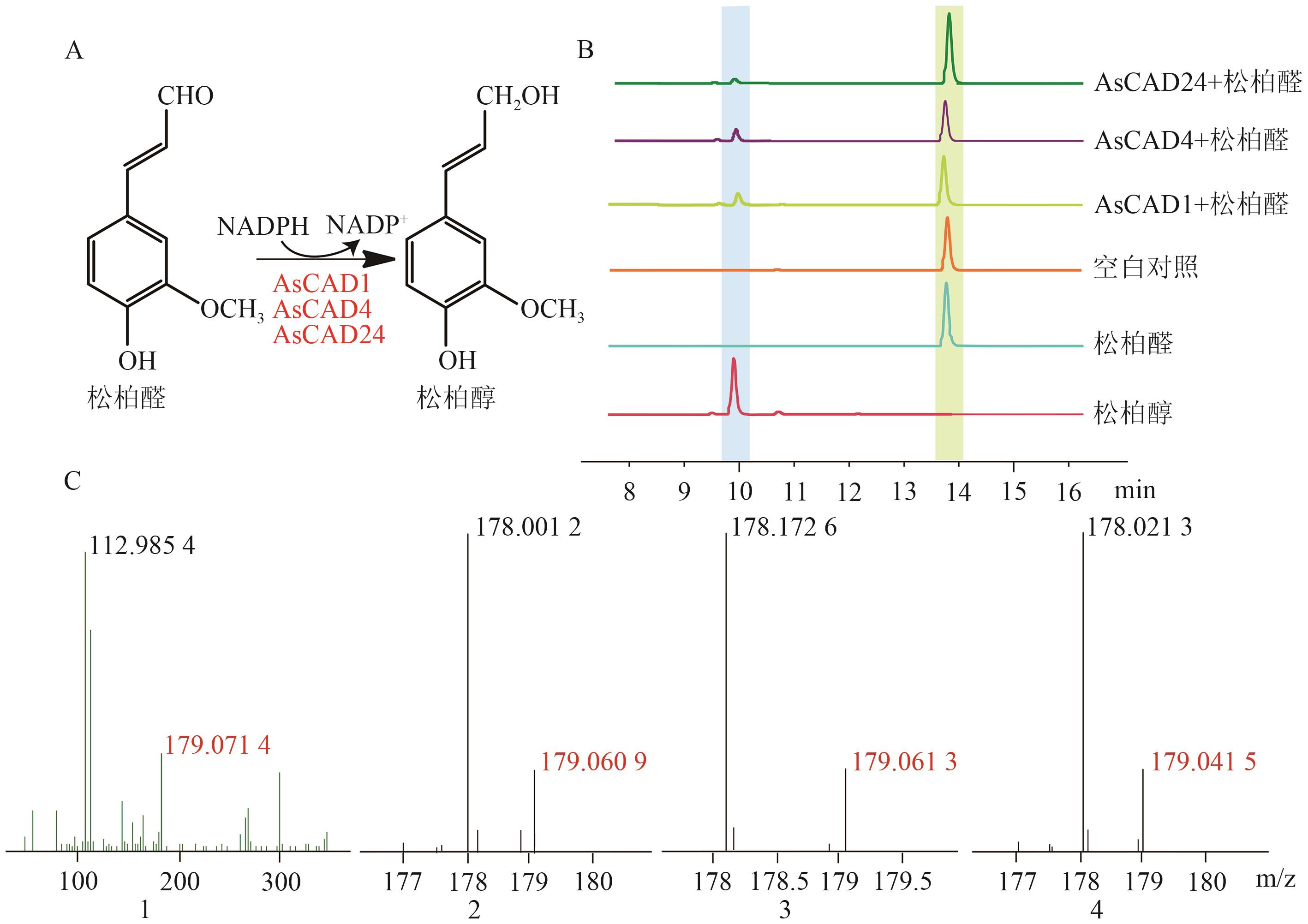
图5 AsCAD1、AsCAD4和AsCAD24催化松柏醛的酶促反应及产物分析A: AsCAD1、AsCAD4和AsCAD24催化松柏醛的还原反应;B: HPLC检测酶促反应混合物及空白对照产物;C: LC-MS检测标准品及酶促反应混合物的分子量(1、2、3、4依次为标准品、AsCAD1、AsCAD4和AsCAD24的LC-MS结果)
Fig. 5 Enzymatic reaction and product analysis of coniferaldehyde catalyzed by AsCAD1, AsCAD4 and AsCAD24A: AsCAD1, AsCAD4 and AsCAD24 catalyze the reduction reaction of coniferaldehyde. B: HPLC is used to detect the products of enzymatic reaction mixtures and blank controls. C: LC-MS is used to detect the molecular weights of standards and enzymatic reaction mixtures (1, 2, 3, and 4 are the LC-MS results of standards, AsCAD1, AsCAD4 and AsCAD24, respectively)

图6 AsCAD1、AsCAD4和AsCAD24催化咖啡醛的酶促反应及产物分析A: AsCAD1、AsCAD4和AsCAD24催化咖啡醛的还原反应;B: HPLC检测酶促反应混合物及空白对照产物;C:LC-MS检测标准品及酶促反应混合物的分子量(1、2、3、4依次为标准品、AsCAD1、AsCAD4和AsCAD24的LC-MS结果)
Fig. 6 Enzymatic reaction and product analysis of caffealdehyde catalyzed by AsCAD1, AsCAD4and AsCAD24A: Reduction reaction of caffealdehyde catalyzed by AsCAD1, AsCAD4 and AsCAD24. B: Detection of enzymatic reaction mixture and blank control product by HPLC. C: Detection of molecular weights of standard and enzymatic reaction mixture by LC-MS (1,2,3,4 are the LC-MS results of standard, AsCAD1, AsCAD4 and AsCAD24, respectively)
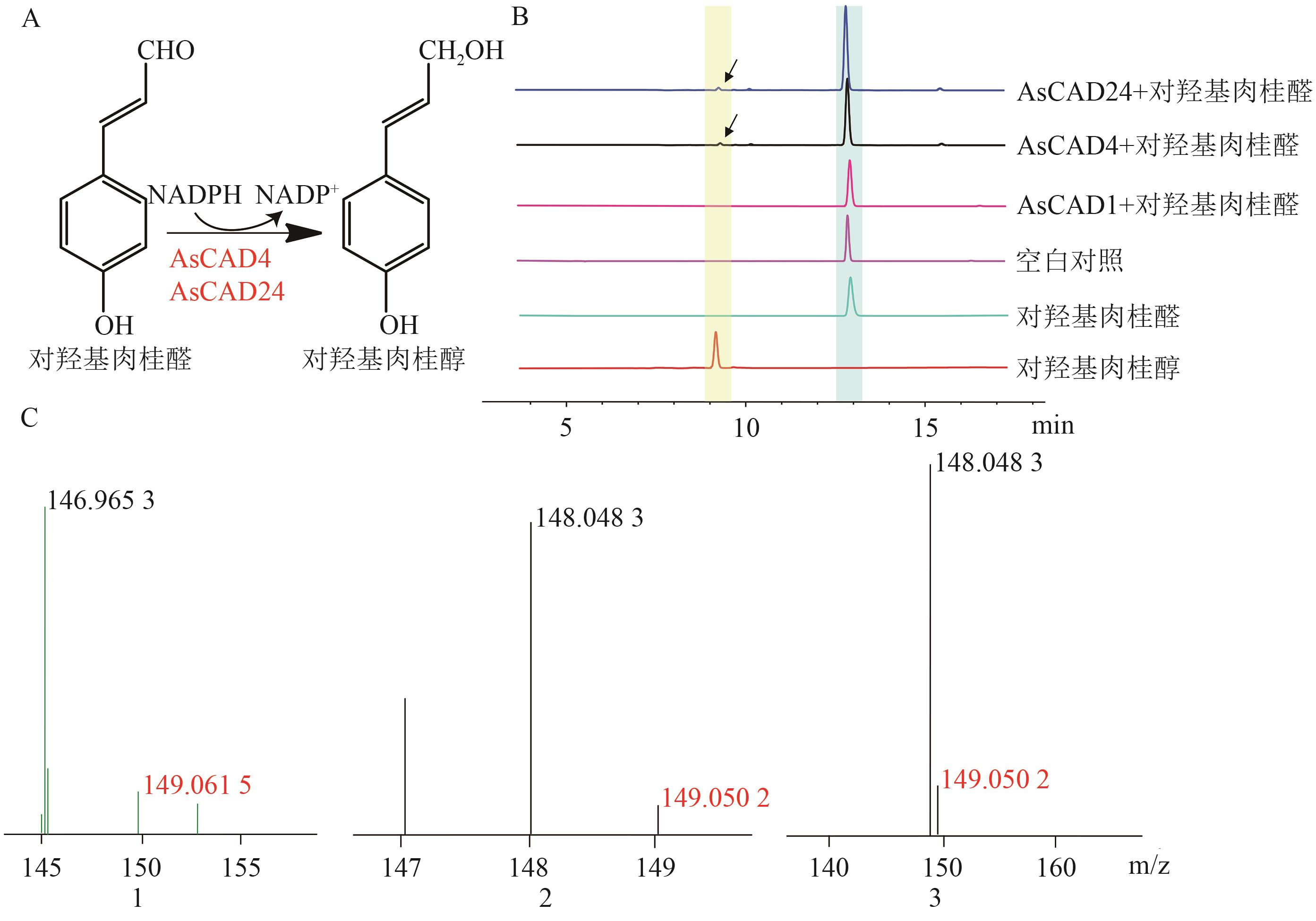
图7 AsCAD1、AsCAD4和AsCAD24催化对羟基肉桂醛的酶促反应及产物分析A: AsCAD4和AsCAD24催化对羟基肉桂醛的还原反应;B: HPLC检测酶促反应混合物及空白对照产物;C:LC-MS检测标准品及酶促反应混合物的分子量(1、2、3依次为标准品、AsCAD4和AsCAD24的LC-MS结果)
Fig. 7 Enzymatic reaction and product analysis of p-coumaraldehyde catalyzed by AsCAD1, AsCAD4 and AsCAD24A: Reduction reaction of p-coumaraldehyde catalysed by AsCAD1, AsCAD4 and AsCAD24. B: Detection of enzymatic reaction mixture and blank control product by HPLC. C: Detection of molecular weights of standard and enzymatic reaction mixture by LC-MS (1,2,3 are the LC-MS results of standard, AsCAD4 and AsCAD24, respectively)

图 8 AsCAD1、AsCAD4和AsCAD24催化芥子醛的酶促反应及产物分析A: AsCAD4和AsCAD24催化芥子醛的还原反应;B: HPLC检测酶促反应混合物及空白对照产物;C: LC-MS检测标准品及酶促反应混合物的分子量(1、2、3依次为标准品、AsCAD4和AsCAD24的LC-MS结果)
Fig. 8 Enzymatic reaction and product analysis of sinapaldehyde catalyzed by AsCAD1, AsCAD4 and AsCAD24A: Reduction reaction of sinapaldehyde catalysed by AsCAD1, AsCAD4 and AsCAD24. B: Detection of enzymatic reaction mixture and blank control product by HPLC. C: Detection of molecular weights of standard and enzymatic reaction mixture by LC-MS (1,2,3 are the LC-MS results of standard, AsCAD4 and AsCAD24, respectively)
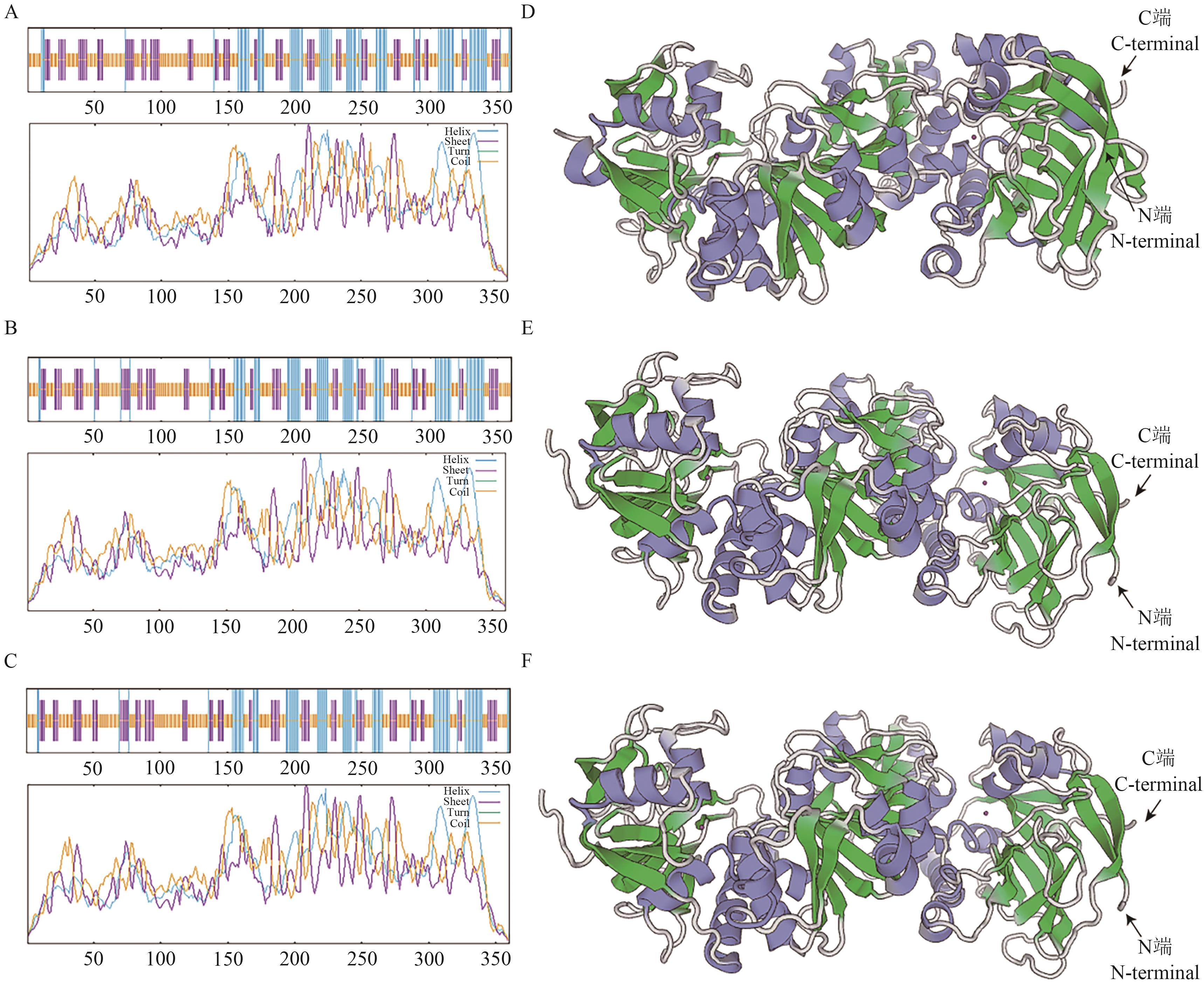
图 9 三个AsCAD蛋白的二级和三级结构预测A-C: AsCAD1、AsCAD4和AsCAD24蛋白二级结构预测;D-F: AsCAD1、AsCAD4和AsCAD24蛋白三级结构预测(紫色:螺旋;绿色:延伸链;银白色:无规则卷曲)
Fig. 9 Secondary and tertiary structure prediction of three AsCAD proteinsA-C: Predicted secondary structures of AsCAD1, AsCAD4 and AsCAD24. D-F: Predicted tertiary structure of AsCAD1, AsCAD4 and AsCAD24 (Purple: Spiral; green: extended chain; silver white: irregular curl)
| 1 | 栗孟飞, 刘学周, 魏建和, 等. 基于生物量、活性物质积累和抗氧化能力的当归高海拔种植区域选择 [J]. 中草药, 2020, 51(2): 474-481. |
| Li MF, Liu XZ, Wei JH, et al. Selection of high altitude planting area of Angelica sinensis based on biomass, bioactive compounds accumulation and antioxidant capacity [J]. Chin Tradit Herb Drugs, 2020, 51(2): 474-481. | |
| 2 | 刘方舟,李园白,王静,等. 当归药材道地性系统评价与分析 [J]. 世界科学技术中医药现代化,2018, 20 (9):1531-1539. |
| Liu FZ, Li YB, Wang J, et al. Systematic evaluation and analysis on Dao-di Herbs Angelica Sinensis [J]. World Science and Technology/Modemization of Traditional Chinese Medicine and Materia Medica J. 2018, 20, (9):1531-1539. | |
| 3 | 马依林, 张虹. 《中华人民共和国药典》中含当归方剂的组方规律 [J]. 中医学报, 2021, 36(12): 2708-2712. |
| Ma YL, Zhang H. Prescription composition principles of Danggui (Radix angelicae sinensis) contained ones in Chinese pharmacopoeia [J]. Acta Chin Med, 2021, 36(12): 2708-2712. | |
| 4 | Yao WL, Zhang L, Hua YL, et al. The investigation of anti-inflammatory activity of volatile oil of Angelica sinensis by plasma metabolomics approach [J]. Int Immunopharmacol, 2015, 29(2): 269-277. |
| 5 | Zhou WJ, Wang S, Hu Z, et al. Angelica sinensis polysaccharides promotes apoptosis in human breast cancer cells via CREB-regulated caspase-3 activation [J]. Biochem Biophys Res Commun, 2015, 467(3): 562-569. |
| 6 | Ma JP, Guo ZB, Jin L, et al. Phytochemical progress made in investigations of Angelica sinensis (Oliv.) Diels [J]. Chin J Nat Med, 2015, 13(4): 241-249. |
| 7 | Wei WL, Zeng R, Gu CM, et al. Angelica sinensis in China-a review of botanical profile, ethnopharmacology, phytochemistry and chemical analysis [J]. J Ethnopharmacol, 2016, 190: 116-141. |
| 8 | 栗孟飞, 康天兰, 晋玲, 等. 当归抽薹开花及其调控途径研究进展 [J]. 中草药, 2020, 51(22): 5894-5899. |
| Li MF, Kang TL, Jin L, et al. Research progress on bolting and flowering of Angelica sinensis and regulation pathways [J]. Chin Tradit Herb Drugs, 2020, 51(22): 5894-5899. | |
| 9 | 梁婕, 王辉, 睢宁. 伞形科根茎类药用植物早期抽薹的研究进展 [J]. 中国农学通报, 2022, 38(13): 90-95. |
| Liang J, Wang H, Sui N. Early bolting of rhizome medicinal plants in Umbelliferae: research progress [J]. Chin Agric Sci Bull, 2022, 38(13): 90-95. | |
| 10 | 马艳春, 吴文轩, 胡建辉, 等. 当归的化学成分及药理作用研究进展 [J]. 中医药学报, 2022, 50(1): 111-114. |
| Ma YC, Wu WX, Hu JH, et al. Research progress on chemical constituents and pharmacological effects of Angelica sinensis [J]. Acta Chin Med Pharmacol, 2022, 50(1): 111-114. | |
| 11 | 管西芹, 毛近隆, 闫滨, 等. 当归不同提取液中阿魏酸、咖啡酸含量及抗氧化作用的比较研究 [J]. 天然产物研究与开发, 2018, 30(12): 2033-2038. |
| Guan XQ, Mao JL, Yan B, et al. A comparative study among ferulic acid, caffeic acid content and antioxidation in different extracts of Angelica sinensis [J]. Nat Prod Res Dev, 2018, 30(12): 2033-2038. | |
| 12 | Ono K, Hirohata M, Yamada M. Ferulic acid destabilizes preformed beta-amyloid fibrils in vitro [J]. Biochem Biophys Res Commun, 2005, 336(2): 444-449. |
| 13 | Li ML, Cui XW, Jin L, et al. Bolting reduces ferulic acid and flavonoid biosynthesis and induces root lignification in Angelica sinensis [J]. Plant Physiol Biochem, 2022, 170: 171-179. |
| 14 | Yuan CX, Li LS, Zhou PH, et al. Decoding the root lignification mechanism of Angelica sinensis through genome-wide methylation analysis [J]. J Exp Bot, 2024: erae392. |
| 15 | Huang LQ, Jin L. Suitable technology for production and processing of Angelica Sinensis [J]. Pharmaceutical Science and Technology Press: Beijing, China, 2018: 1-14. |
| 16 | 张真, 刘学周, 包亚军, 等. 基于生物量、活性物质积累和抗氧化能力的当归种植茬口选择 [J]. 甘肃农业大学学报, 2018, 53(6): 82-89. |
| Zhang Z, Liu XZ, Bao YJ, et al. Selection of cropping rotations of Angelicasinensisbased on biomass, bioactive compounds accumulation and antioxidant capacity [J]. J Gansu Agric Univ, 2018, 53(6): 82-89. | |
| 17 | Miedes E, Vanholme R, Boerjan W, et al. The role of the secondary cell wall in plant resistance to pathogens [J]. Front Plant Sci, 2014, 5: 358. |
| 18 | Boerjan W, Ralph J, Baucher M. Lignin biosynthesis [J]. Annu Rev Plant Biol, 2003, 54: 519-546. |
| 19 | Bonawitz ND, Chapple C. The genetics of lignin biosynthesis: connecting genotype to phenotype [J]. Annu Rev Genet, 2010, 44: 337-363. |
| 20 | Baucher M, Chabbert B, Pilate G, et al. Red xylem and higher lignin extractability by down-regulating a cinnamyl alcohol dehydrogenase in poplar [J]. Plant Physiol, 1996, 112(4): 1479-1490. |
| 21 | Halpin C, Knight ME, Grima-Pettenati J, et al. Purification and characterization of cinnamyl alcohol dehydrogenase from tobacco stems [J]. Plant Physiol, 1992, 98(1): 12-16. |
| 22 | Ma QH. Functional analysis of a cinnamyl alcohol dehydrogenase involved in lignin biosynthesis in wheat [J]. J Exp Bot, 2010, 61(10): 2735-2744. |
| 23 | Sibout R, Eudes A, Pollet B, et al. Expression pattern of two paralogs encoding cinnamyl alcohol dehydrogenases in Arabidopsis. Isolation and characterization of the corresponding mutants [J]. Plant Physiol, 2003, 132(2): 848-860. |
| 24 | Zhang KW, Qian Q, Huang ZJ, et al. GOLD HULL AND INTERNODE2 encodes a primarily multifunctional cinnamyl-alcohol dehydrogenase in rice [J]. Plant Physiol, 2006, 140(3): 972-983. |
| 25 | Chao N, Liu SX, Liu BM, et al. Molecular cloning and functional analysis of nine cinnamyl alcohol dehydrogenase family members in Populus tomentosa [J]. Planta, 2014, 240(5): 1097-1112. |
| 26 | Porter S, Sederoff RR. Purification, characterization, and cloning of cinnamyl alcohol dehydrogenase in loblolly pine (Pinus taeda L.) [J]. Plant Physiol, 1992, 98(4): 1364-1371. |
| 27 | Lee CJ, Kim SE, Park SU, et al. Tuberous roots of transgenic sweetpotato overexpressing IbCAD1 have enhanced low-temperature storage phenotypes [J]. Plant Physiol Biochem, 2021, 166: 549-557. |
| 28 | Park HL, Kim TL, Bhoo SH, et al. Biochemical characterization of the rice cinnamyl alcohol dehydrogenase gene family [J]. Molecules, 2018, 23(10): 2659. |
| 29 | Pan HY, Zhou R, Louie GV, et al. Structural studies of cinnamoyl-CoA reductase and cinnamyl-alcohol dehydrogenase, key enzymes of monolignol biosynthesis [J]. Plant Cell, 2014, 26(9): 3709-3727. |
| 30 | Vasupalli N, Hou D, Singh RM, et al. Homo- and hetero-dimers of CAD enzymes regulate lignification and abiotic stress response in moso bamboo [J]. Int J Mol Sci, 2021, 22(23): 12917. |
| 31 | Baghdady A, Blervacq AS, Jouanin L, et al. Eucalyptus gunnii CCR and CAD2 promoters are active in lignifying cells during primary and secondary xylem formation in Arabidopsis thaliana [J]. Plant Physiol Biochem, 2006, 44(11-12): 674-683. |
| 32 | Kim SJ, Kim MR, Bedgar DL, et al. Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis [J]. Proc Natl Acad Sci USA, 2004, 101(6): 1455-1460. |
| 33 | 朋冬琴, 罗蜜蜜, 郭欣慰, 等. 当归实时荧光定量PCR内参基因筛选 [J]. 中草药, 2024, 55(1): 269-278. |
| Peng DQ, Luo MM, Guo XW, et al. Selection of reference genes for quantitative real-time PCR analysis in Angelica sinensis [J]. Chin Tradit Herb Drugs, 2024, 55(1): 269-278. | |
| 34 | Rong W, Luo MY, Shan TL, et al. A wheat cinnamyl alcohol dehydrogenase TaCAD12 contributes to host resistance to the sharp eyespot disease [J]. Front Plant Sci, 2016, 7: 1723. |
| 35 | Tobias CM, Chow EK. Structure of the cinnamyl-alcohol dehydrogenase gene family in rice and promoter activity of a member associated with lignification [J]. Planta, 2005, 220(5): 678-688. |
| 36 | Li X, Ma DM, Chen JL, et al. Biochemical characterization and identification of a cinnamyl alcohol dehydrogenase from Artemisia annua [J]. Plant Sci, 2012, 193/194: 85-95. |
| 37 | Barakat A, Bagniewska-Zadworna A, Choi A, et al. The cinnamyl alcohol dehydrogenase gene family in Populus: phylogeny, organization, and expression [J]. BMC Plant Biol, 2009, 9: 26. |
| 38 | Kim SJ, Kim KW, Cho MH, et al. Expression of cinnamyl alcohol dehydrogenases and their putative homologues during Arabidopsis thaliana growth and development: lessons for database annotations? [J]. Phytochemistry, 2007, 68(14): 1957-1974. |
| 39 | Chen F, Tobimatsu Y, Havkin-Frenkel D, et al. A polymer of caffeyl alcohol in plant seeds [J]. Proc Natl Acad Sci USA, 2012, 109(5): 1772-1777. |
| 40 | Tobimatsu Y, Chen F, Nakashima J, et al. Coexistence but independent biosynthesis of catechyl and guaiacyl/syringyl lignin polymers in seed Coats [J]. Plant Cell, 2013, 25(7): 2587-2600. |
| 41 | Chen F, Tobimatsu Y, Jackson L, et al. Novel seed coat lignins in the Cactaceae: structure, distribution and implications for the evolution of lignin diversity [J]. Plant J, 2013, 73(2): 201-211. |
| 42 | Youn B, Camacho R, Moinuddin SGA, et al. Crystal structures and catalytic mechanism of the Arabidopsis cinnamyl alcohol dehydrogenases AtCAD5 and AtCAD4 [J]. Org Biomol Chem, 2006, 4(9): 1687-1697. |
| 43 | 王贺萍, 孙震, 刘雨辰, 等. 蒙古冰草肉桂醇脱氢酶基因序列鉴定及功能分析 [J]. 植物学报, 2024, 59(2): 204-216. |
| Wang HP, Sun Z, Liu YC, et al. Sequence identification and functional analysis of cinnamyl alcohol dehydrogenase gene from Agropyron mongolicum [J]. Chin Bull Bot, 2024, 59(2): 204-216. | |
| 44 | Bomati EK, Noel JP. Structural and kinetic basis for substrate selectivity in Populus tremuloides sinapyl alcohol dehydrogenase [J]. Plant Cell, 2005, 17(5): 1598-1611. |
| 45 | Yang M, Fehl C, Lees KV, et al. Functional and informatics analysis enables glycosyltransferase activity prediction [J]. Nat Chem Biol, 2018, 14(12): 1109-1117. |
| 46 | 宋希茜. 超积累型东南景天SaCAD基因的克隆及其功能分析 [D]. 北京: 中国林业科学研究院, 2016. |
| Song XQ. Cloning and functional analysis of SaCAD gene of hyperaccumulator Sedum alfredii [D]. Beijing: Chinese Academy of Forestry, 2016. | |
| 47 | Ponniah SK, Shang ZH, Akbudak MA, et al. Down-regulation of hydroxycinnamoyl CoA: shikimate hydroxycinnamoyl transferase, cinnamoyl CoA reductase, and cinnamyl alcohol dehydrogenase leads to lignin reduction in rice (Oryza sativa L. ssp. Japonica cv. nipponbare) [J]. Plant Biotechnol Rep, 2017, 11(1): 17-27. |
| 48 | Xun HW, Qian XY, Wang M, et al. Overexpression of a cinnamyl alcohol dehydrogenase-coding gene, GsCAD1, from wild soybean enhances resistance to soybean mosaic virus [J]. Int J Mol Sci, 2022, 23(23): 15206. |
| [1] | 马天意, 许家佳, 路文婧, 吴艳, 沙伟, 张梅娟, 彭疑芳. ‘金小童’大白菜BrcGASA3基因在盐碱胁迫下的表达分析及抗性鉴定[J]. 生物技术通报, 2025, 41(2): 127-138. |
| [2] | 许圆梦, 毛娇, 王梦瑶, 王数, 任江陵, 刘宇涵, 刘思辰, 乔治军, 王瑞云, 曹晓宁. 糜子PmDEP1和PmEP3基因的克隆与表达特征分析[J]. 生物技术通报, 2025, 41(2): 150-162. |
| [3] | 贾子健, 王宝强, 陈立飞, 王义真, 魏小红, 赵颖. 响应NO的藜麦CHX基因家族在盐碱胁迫下的表达模式[J]. 生物技术通报, 2025, 41(2): 163-174. |
| [4] | 钱政毅, 吴绍芳, 曹舒怡, 宋雅欣, 潘鑫峰, 李兆伟, 范凯. 睡莲NAC转录因子的鉴定及其表达分析[J]. 生物技术通报, 2025, 41(2): 234-247. |
| [5] | 李禹欣, 李苗, 杜晓芬, 韩康妮, 连世超, 王军. 谷子SiSAP基因家族的鉴定与表达分析[J]. 生物技术通报, 2025, 41(1): 143-156. |
| [6] | 孔青洋, 张晓龙, 李娜, 张晨洁, 张雪云, 于超, 张启翔, 罗乐. 单叶蔷薇GRAS转录因子家族鉴定及表达分析[J]. 生物技术通报, 2025, 41(1): 210-220. |
| [7] | 宋兵芳, 柳宁, 程新艳, 徐晓斌, 田文茂, 高悦, 毕阳, 王毅. 马铃薯G6PDH基因家族鉴定及其在损伤块茎的表达分析[J]. 生物技术通报, 2024, 40(9): 104-112. |
| [8] | 吴慧琴, 王延宏, 刘涵, 司政, 刘雪晴, 王静, 阳宜, 成妍. 辣椒UGT基因家族的鉴定及表达分析[J]. 生物技术通报, 2024, 40(9): 198-211. |
| [9] | 满全财, 孟姿诺, 李伟, 蔡心汝, 苏润东, 付长青, 高顺娟, 崔江慧. 马铃薯AQP基因家族鉴定及表达分析[J]. 生物技术通报, 2024, 40(9): 51-63. |
| [10] | 申鹏, 高雅彬, 丁红. 马铃薯SAT基因家族的鉴定和表达分析[J]. 生物技术通报, 2024, 40(9): 64-73. |
| [11] | 李亦君, 杨小贝, 夏琳, 罗朝鹏, 徐馨, 杨军, 宁黔冀, 武明珠. 烟草NtPRR37基因克隆及功能分析[J]. 生物技术通报, 2024, 40(8): 221-231. |
| [12] | 李雨晴, 吴楠, 罗建让. 卵叶牡丹花色苷合成相关基因bHLH的克隆与功能分析[J]. 生物技术通报, 2024, 40(8): 174-185. |
| [13] | 崔原瑗, 王昭懿, 白双宇, 任毓昭, 豆飞飞, 刘彩霞, 刘凤楼, 王掌军, 李清峰. 大麦非特异性磷脂酶C基因家族全基因组鉴定及苗期胁迫表达分析[J]. 生物技术通报, 2024, 40(8): 74-82. |
| [14] | 杨巍, 赵丽芬, 唐兵, 周麟笔, 杨娟, 莫传园, 张宝会, 李飞, 阮松林, 邓英. 芥菜SRO基因家族全基因组鉴定与表达分析[J]. 生物技术通报, 2024, 40(8): 129-141. |
| [15] | 周冉, 王兴平, 李彦霞, 罗仍卓么. 金黄色葡萄球菌型乳房炎奶牛乳腺组织的lncRNA差异表达分析[J]. 生物技术通报, 2024, 40(8): 320-328. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||