








生物技术通报 ›› 2026, Vol. 42 ›› Issue (4): 129-140.doi: 10.13560/j.cnki.biotech.bull.1985.2025-1008
• 研究报告 • 上一篇
王玉昆1,2,3( ), 原远2,3, 王斌2,3, 朱云娜2,3, 任晓强2,3, 任飞2,3(
), 原远2,3, 王斌2,3, 朱云娜2,3, 任晓强2,3, 任飞2,3( ), 叶红2,3(
), 叶红2,3( )
)
收稿日期:2025-09-22
出版日期:2026-02-09
发布日期:2026-02-09
通讯作者:
任飞,女,博士,讲师,研究方向 :药用植物分子生物学;E-mail: ren_faye_sgu@163.com作者简介:王玉昆,男,博士,副教授,研究方向 :园艺植物分子生物学;E-mail: wangyu_kun1@163.com
基金资助:
WANG Yu-kun1,2,3( ), YUAN Yuan2,3, WANG Bin2,3, ZHU Yun-na2,3, REN Xiao-qiang2,3, REN Fei2,3(
), YUAN Yuan2,3, WANG Bin2,3, ZHU Yun-na2,3, REN Xiao-qiang2,3, REN Fei2,3( ), YE Hong2,3(
), YE Hong2,3( )
)
Received:2025-09-22
Published:2026-02-09
Online:2026-02-09
摘要:
目的 解析紫苏(Perilla frutescens)QO10和QS5种子α-亚麻酸(α-linolenic acid, ALA)含量差异,挖掘ALA合成及调控相关基因。为进一步创制高ALA含量的紫苏新品种提供遗传资源。 方法 采用形态学指标检测、转录组学和脂质代谢组学技术,对紫苏QO10和QS5种子形态、差异表达基因(differentially expressed genes, DEGs)和脂质代谢物谱进行系统分析。 结果 QS5种子千粒重显著高于QO10。种皮色差分析表明QS5种皮颜色比QO10更亮,二者在种皮颜色上极易区分。脂质代谢组分析结果表明,QO10和QS5种子中脂质代谢物种类和相对含量具有明显差异,QS5种子中硬脂酸(stearic acid, SA)和ALA含量高于QO10。转录组分析结果显示,188个差异表达基因(DEGs)富集到6个与脂肪酸(fatty acid, FA)代谢相关的GO条目和5个KEGG通路。依据转录组‒脂质代谢组联合分析结果,最终筛选出15个关键酶及蛋白编码基因,包括直接参与ALA生物合成调控的ACSL、FABP、FAD2、ENR、KAR和KAS,以及间接参与ALA生物合成调控的脂肪酶(lipase)编码基因、LCAT3、FAR、SCL及HMGCR。此外,转录组分析显示38个WRKY转录因子(transcription factor, TF)和26个MYB TFs差异表达,并筛选了差异表达倍数最大的2个WRKY和2个MYB TFs作为候选TFs。最后,通过RT-qPCR验证10个与ALA生物合成调控相关的候选基因,定量结果与转录组测序结果一致。 结论 紫苏QO10与QS5种子在形态学上差异显著,且QS5含有更高的SA和ALA。筛选出15个酶编码基因和4个TFs,这些基因在QO10和QS5种子中的差异表达是造成ALA含量差异的可能原因。
王玉昆, 原远, 王斌, 朱云娜, 任晓强, 任飞, 叶红. 转录组和脂质代谢组联合分析不同紫苏α-亚麻酸合成调控差异[J]. 生物技术通报, 2026, 42(4): 129-140.
WANG Yu-kun, YUAN Yuan, WANG Bin, ZHU Yun-na, REN Xiao-qiang, REN Fei, YE Hong. Integrated Analysis of Transcriptome and Lipid Metabolome Reveals the Differences in α-Linolenic Acid Synthesis Regulation in Different Perilla frutescens[J]. Biotechnology Bulletin, 2026, 42(4): 129-140.
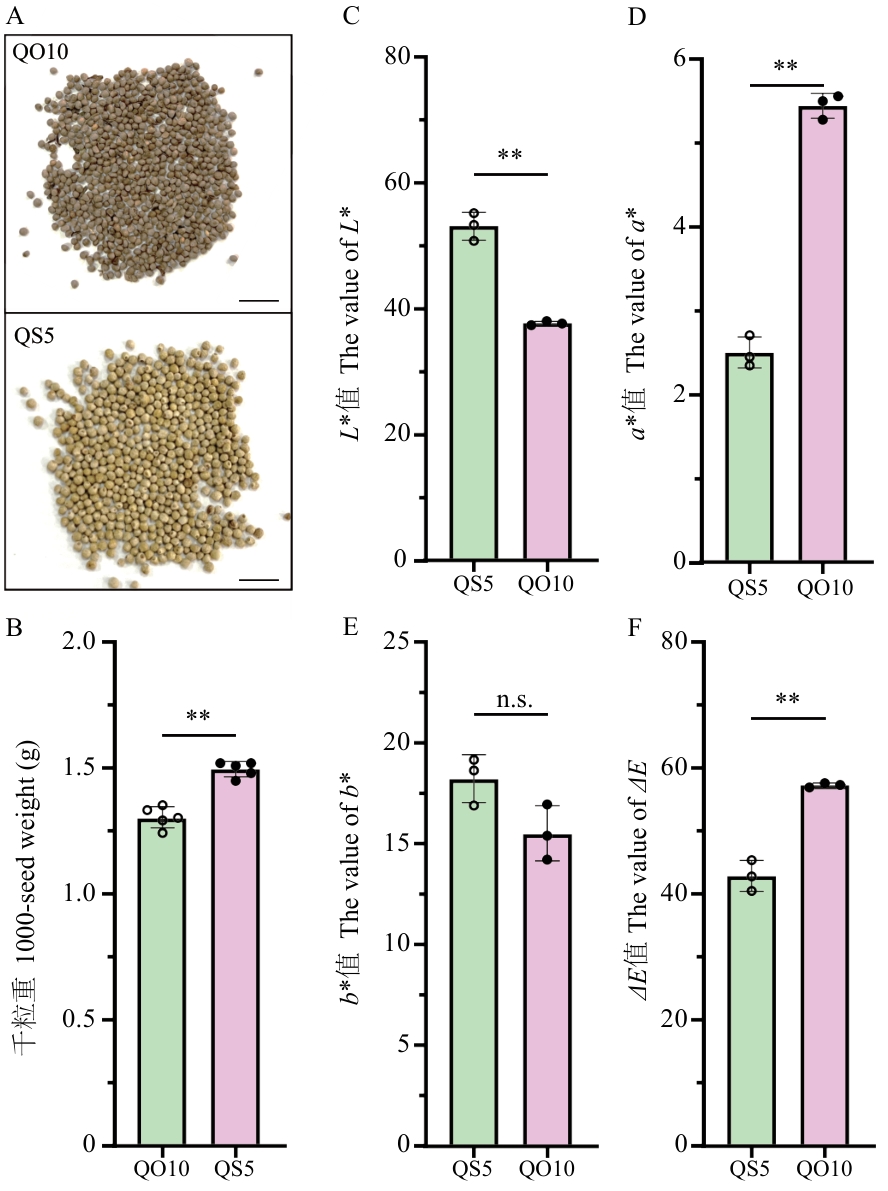
图1 紫苏QO10和QS5种子的形态学观察A:QO10和QS5种子形态;B:QO10和QS5种子千粒重;C‒F:QO10和QS5种子种皮色差参数测定;L*值表示颜色的明亮程度;a*值表示颜色在“红色‒绿色”轴上的偏向;b*值表示颜色在“黄色‒蓝色”轴上的偏向;ΔE值表示综合L*、a*、b* 3个维度的差异,计算得出的总色差值;**P<0.01;n.s.:无显著差异;比例尺=1 cm
Fig. 1 Morphological observation of perilla (Perilla frutescens) QO10 and QS5 seedsA: Morphology of QO10 and QS5 seeds; B: 1 000-seed weight of QO10 and QS5 seeds; C‒F: determination of color difference parameters of seed coats of QO10 and QS5 seeds. The L* value indicates the lightness of a color. The a* value indicates the color’s bias on the “red-green” axis. The b* value indicates the color’s bias on the “yellow-blue” axis. The ΔE value indicates the total color difference value calculated by integrating the differences in the three dimensions of L*, a*, and b*. ** P<0.01; n.s..: no significant difference. Scale bar=1 cm
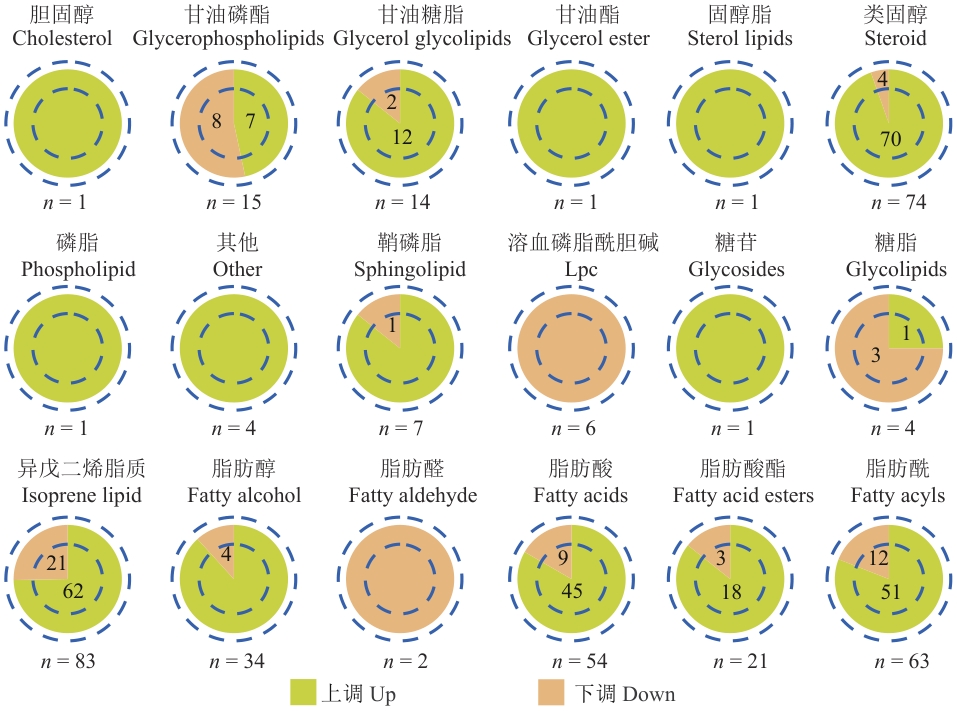
图2 紫苏QO10和QS5种子中差异脂质代谢物的种类、数量及积累模式
Fig. 2 Types, quantities, and accumulation patterns of differential lipid metabolites in perilla QO10 and QS5 seeds
| 样本 Samples | 总读数 Total reads | 干净读数 Clean reads | 干净碱基 Clean bases | GC含量 GC content (%) | Q30百分比Q30 percentage (%) | 比对读数 Mapped reads | 特异比对读数 Unique mapped reads |
|---|---|---|---|---|---|---|---|
| Q010-1 | 43 192 794 | 21 596 397 | 6 460 553 535 | 47.69 | 94.72 | 41 275 128 (95.56%) | 38 960 150 (90.20%) |
| Q010-2 | 49 973 860 | 24 986 930 | 7 472 777 948 | 47.50 | 94.35 | 47 022 281 (94.09%) | 44 202 523 (88.45%) |
| Q010-3 | 41 845 218 | 20 922 609 | 6 256 919 302 | 47.65 | 94.85 | 39 954 021 (95.48%) | 37 653 099 (89.98%) |
| QS5-1 | 38 146 772 | 19 073 386 | 5 698 842 059 | 49.37 | 94.56 | 34 751 710 (91.10%) | 30 637 108 (88.16%) |
| QS5-2 | 36 792 684 | 18 396 342 | 5 494 890 960 | 49.46 | 94.45 | 34 287 102 (93.19%) | 30 669 813 (89.45%) |
| QS5-3 | 36 323 210 | 18 161 605 | 5 424 955 610 | 49.49 | 94.73 | 34 314 537 (94.47%) | 30 502 192 (88.89%) |
表2 转录组测序数据统计
Table 2 Transcriptome sequencing data statistics
| 样本 Samples | 总读数 Total reads | 干净读数 Clean reads | 干净碱基 Clean bases | GC含量 GC content (%) | Q30百分比Q30 percentage (%) | 比对读数 Mapped reads | 特异比对读数 Unique mapped reads |
|---|---|---|---|---|---|---|---|
| Q010-1 | 43 192 794 | 21 596 397 | 6 460 553 535 | 47.69 | 94.72 | 41 275 128 (95.56%) | 38 960 150 (90.20%) |
| Q010-2 | 49 973 860 | 24 986 930 | 7 472 777 948 | 47.50 | 94.35 | 47 022 281 (94.09%) | 44 202 523 (88.45%) |
| Q010-3 | 41 845 218 | 20 922 609 | 6 256 919 302 | 47.65 | 94.85 | 39 954 021 (95.48%) | 37 653 099 (89.98%) |
| QS5-1 | 38 146 772 | 19 073 386 | 5 698 842 059 | 49.37 | 94.56 | 34 751 710 (91.10%) | 30 637 108 (88.16%) |
| QS5-2 | 36 792 684 | 18 396 342 | 5 494 890 960 | 49.46 | 94.45 | 34 287 102 (93.19%) | 30 669 813 (89.45%) |
| QS5-3 | 36 323 210 | 18 161 605 | 5 424 955 610 | 49.49 | 94.73 | 34 314 537 (94.47%) | 30 502 192 (88.89%) |
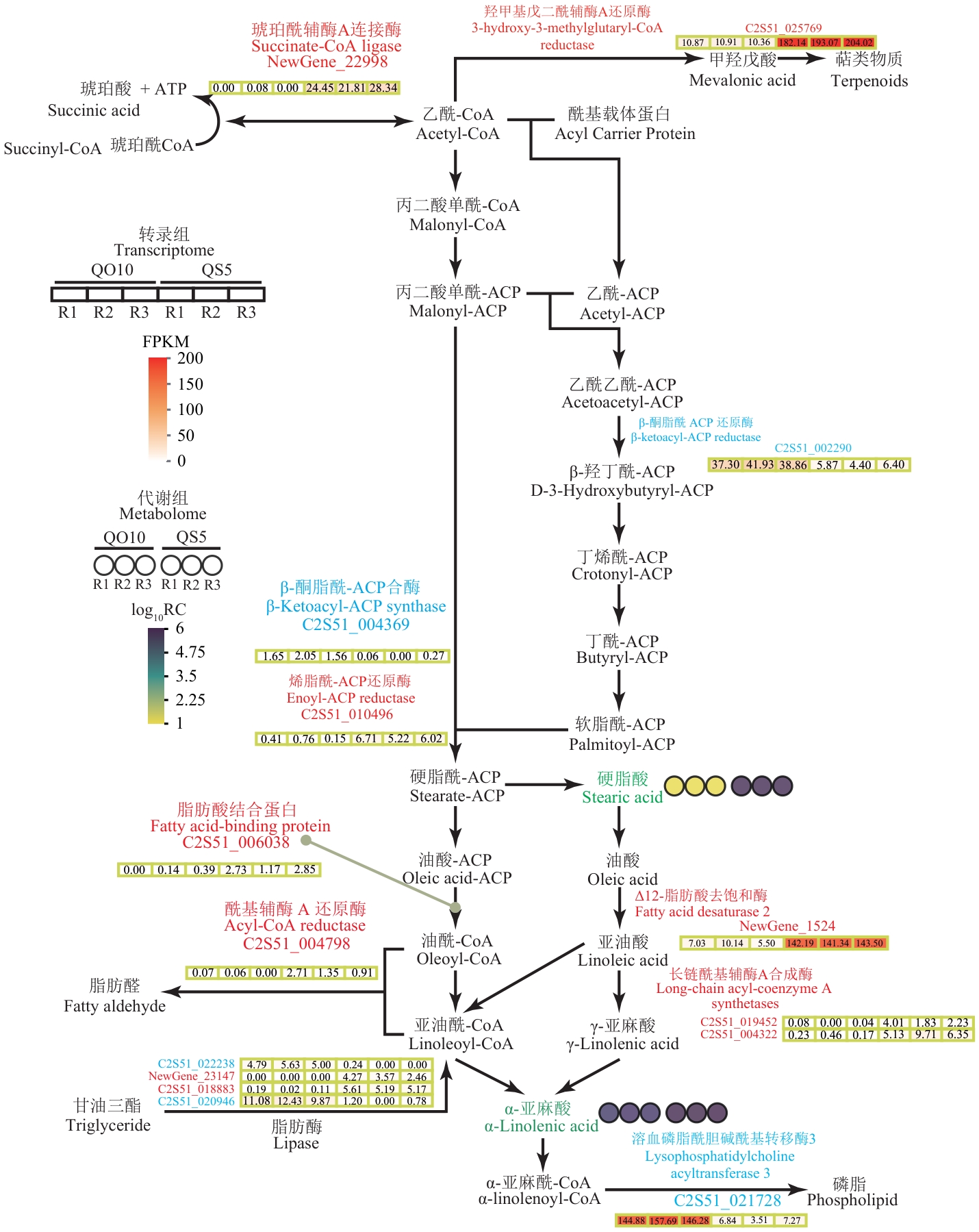
图7 紫苏种子ALA合成代谢转录组‒代谢组映射通路红色字体表示上调,蓝色字体表示下调。转录组热图表示基因在不同样品中的表达水平(FPKM),代谢组热图表示代谢物在不同样品中的积累量。R1‒R3表示生物学重复
Fig. 7 Joint analysis of transcriptome and metabolome of ALA anabolism in perilla seedsRed font indicates upregulation and blue font indicates downregulation. The transcriptome heatmap represents the expression levels (FPKM) of genes in different samples, and the metabolome heatmap indicates the accumulation of metabolites in different samples. R1‒R3 indicates biological replications
| [1] | Tvrzicka E, Kremmyda LS, Stankova B, et al. Fatty acids as biocompounds: their role in human metabolism, health and disease—a review. Part 1: classification, dietary sources and biological functions [J]. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub, 2011, 155(2): 117-130. |
| [2] | He M, Ding NZ. Plant unsaturated fatty acids: multiple roles in stress response [J]. Front Plant Sci, 2020, 11: 562785. |
| [3] | Baker EJ, Miles EA, Burdge GC, et al. Metabolism and functional effects of plant-derived omega-3 fatty acids in humans [J]. Prog Lipid Res, 2016, 64: 30-56. |
| [4] | Egert S, Baxheinrich A, Lee-Barkey YH, et al. Effects of a hypoenergetic diet rich in α-linolenic acid on fatty acid composition of serum phospholipids in overweight and obese patients with metabolic syndrome [J]. Nutrition, 2018, 49: 74-80. |
| [5] | Li JJ, Gu ZN, Pan Y, et al. Dietary supplementation of α-linolenic acid induced conversion of n-3 LCPUFAs and reduced prostate cancer growth in a mouse model [J]. Lipids Health Dis, 2017, 16(1): 136. |
| [6] | Litwiniuk A, Domańska A, Chmielowska M, et al. The effects of alpha-linolenic acid on the secretory activity of astrocytes and β amyloid-associated neurodegeneration in differentiated SH-SY5Y cells: alpha-linolenic acid protects the SH-SY5Y cells against β amyloid toxicity [J]. Oxid Med Cell Longev, 2020, 2020(1): 8908901. |
| [7] | Tang YH, Jiang Y, Meng JS, et al. A brief review of physiological roles, plant resources, synthesis, purification and oxidative stability of alpha-linolenic acid [J]. Emir J Food Agric, 2018, 30(5):341-356. |
| [8] | Potu RB, Lu H, Adeola O, et al. Metabolic markers in Ossabaw pigs fed high fat diets enriched in regular or low α-linolenic acid soy oil [J]. Nutr Metab, 2013, 10(1): 27. |
| [9] | Deng QC, Yu X, Xu JQ, et al. Single frequency intake of α-linolenic acid rich phytosterol esters attenuates atherosclerosis risk factors in hamsters fed a high fat diet [J]. Lipids Health Dis, 2016, 15: 23. |
| [10] | Heskey CE, Jaceldo-Siegl K, Sabaté J, et al. Adipose tissue α-linolenic acid is inversely associated with insulin resistance in adults [J]. Am J Clin Nutr, 2016, 103(4): 1105-1110. |
| [11] | Sala-Vila A, Guasch-Ferré M, Hu FB, et al. Dietary α-linolenic acid, marine ω-3 fatty acids, and mortality in a population with high fish consumption: findings from the PREvención con DIeta MEDiterránea (PREDIMED) study [J]. J Am Heart Assoc, 2016, 5(1): e002543. |
| [12] | Skilton MR, Pahkala K, Viikari JSA, et al. The association of dietary alpha-linolenic acid with blood pressure and subclinical atherosclerosis in people born small for gestational age: the special turku coronary risk factor intervention project study [J]. J Pediatr, 2015, 166(5): 1252-1257.e2. |
| [13] | Yue H, Qiu B, Jia M, et al. Effects of α-linolenic acid intake on blood lipid profiles: a systematic review and meta-analysis of randomized controlled trials [J]. Crit Rev Food Sci Nutr, 2021, 61(17): 2894-2910. |
| [14] | 徐爱遐, 张博勇, 赵德义, 等. 富含α-亚麻酸植物的研究 [J]. 水土保持学报, 2004(4): 190-192, 199. |
| Xu AX, Zhang BY, Zhao DY, et al. Study on plants rich in α-linolenic acid [J]. J Soil Water Conserv, 2004(4): 190-192, 199. | |
| [15] | 张丽娟, 杨解顺, 殷建忠. 富含α-亚麻酸唇形科植物油的研究进展 [J]. 国外医学: 医学地理分册, 2010, 31(2): 127-129. |
| Zhang LJ, Yang JS, Yin JZ. Research progress of α-linolenic-rich acid labiate oils [J]. Foreign Med Sci Sect Medgeography, 2010, 31(2): 127-129. | |
| [16] | 周雅莉, 黄旭升, 李润植, 等. 食药同源作物紫苏α-亚麻酸生物合成及调控机制的研究 [C]//第二十届中国作物学会学术年会论文集. 长沙, 2023: 322. |
| Zhou YL, Huang XS, Li RZ, et al. Study on the biosynthesis and regulatory mechanism of α-linolenic acid in Perilla frutescens, a crop with homologous food and medicine [C]// Abstracts of the 20th Academic Annual Meeting of the Chinese Crop Science Society. Changsha, 2023: 322. | |
| [17] | 耿鑫鑫, 周讯, 杨凤琳, 等. 紫苏种子中α-亚麻酸积累关键基因的筛选及克隆 [J]. 湖北师范大学学报: 自然科学版, 2025, 45(2): 69-77. |
| Geng XX, Zhou X, Yang FL, et al. Screening and cloning of key genes for α-linolenic acid accumulation in Perilla frutescens seeds [J]. J Hubei Norm Univ Nat Sci, 2025, 45(2): 69-77. | |
| [18] | Zhang YJ, Shen Q, Leng L, et al. Incipient diploidization of the medicinal plant perilla within 10, 000 years [J]. Nat Commun, 2021, 12(1): 5508. |
| [19] | Ye H, Mu JX, Yang T, et al. Integrated transcriptome and metabolome analyses reveal candidate genes associate with phenolic compound biosynthesis in different varieties of Perilla frutescens [J]. Int J Mol Sci, 2025, 26(7): 2841. |
| [20] | 吴端, 王力军, 杨仕梅, 等. 植物种子α-亚麻酸形成及调控机理研究进展 [J]. 植物遗传资源学报, 2020, 21(1): 49-62. |
| Wu D, Wang LJ, Yang SM, et al. Advances on formation and regulation mechanism of α-linolenic acid in seeds [J]. J Plant Genet Resour, 2020, 21(1): 49-62. | |
| [21] | 邓咪咪, 刘宝玲, 王志龙, 等. 大豆硬脂酰-ACP Δ9脱氢酶(GmSAD)基因家族的鉴定及功能分析 [J]. 生物工程学报, 2020, 36(4): 716-731. |
| Deng MM, Liu BL, Wang ZL, et al. Identification and functional analysis of soybean stearoyl-ACP Δ9 desaturase (GmSAD) gene family [J]. Chin J Biotechnol, 2020, 36(4): 716-731. | |
| [22] | Arondel V, Vergnolle C, Tchang F, et al. Bifunctional lipid-transfer: fatty acid-binding proteins in plants [J]. Mol Cell Biochem, 1990, 98(1): 49-56. |
| [23] | Xiao RX, Zou YR, Guo XR, et al. Fatty acid desaturases (FADs) modulate multiple lipid metabolism pathways to improve plant resistance [J]. Mol Biol Rep, 2022, 49(10): 9997-10011. |
| [24] | Buchert R, Tawamie H, Smith C, et al. A peroxisomal disorder of severe intellectual disability, epilepsy, and cataracts due to fatty acyl-CoA reductase 1 deficiency [J]. Am J Hum Genet, 2014, 95(5): 602-610. |
| [25] | Kelly AA, Feussner I. Oil is on the agenda: Lipid turnover in higher plants [J]. Biochim Biophys Acta Mol Cell Biol Lipds, 2016, 1861(9): 1253-1268. |
| [26] | Ou XB, Wang X, Zhao B, et al. Metabolome and transcriptome signatures shed light on the anti-obesity effect of Polygonatum sibiricum [J]. Front Plant Sci, 2023, 14: 1181861. |
| [27] | Studart-Guimarães C, Gibon Y, Frankel N, et al. Identification and characterisation of the alpha and beta subunits of succinyl CoA ligase of tomato [J]. Plant Mol Biol, 2005, 59(5): 781-791. |
| [28] | 郑婷, 魏灵珠, 程建徽, 等. 植物3-羟基-3-甲基戊二酰辅酶A还原酶(HMGR)研究进展 [J]. 植物生理学报, 2022, 58(6): 1037-1044. |
| Zheng T, Wei LZ, Cheng JH, et al. Research progress of 3-hydroxy-3-methylglutaryl-CoA reductase in plants [J]. Plant Physiol J, 2022, 58(6): 1037-1044. | |
| [29] | 惠生娟, 葛丽萍, 王子瑜, 等. 续随子MYB基因家族的鉴定及ElMYB114在油脂合成中的功能分析 [J]. 植物科学学报, 2025, 43(1): 92-101. |
| Hui SJ, Ge LP, Wang ZY, et al. Identification of MYB gene family and functional analysis of ElMYB114 in lipid synthesis of Euphorbia lathyris L [J]. Plant Sci J, 2025, 43(1): 92-101. | |
| [30] | Xie GW, Zhang YX, Xiao S, et al. Molecular mapping of candidate genes in determining red color of perilla leaf [J]. Adv Biotechnol, 2025, 3(1): 7. |
| [31] | Zhang TY, Song C, Song L, et al. RNA sequencing and coexpression analysis reveal key genes involved in α-linolenic acid biosynthesis in Perilla frutescens seed [J]. Int J Mol Sci, 2017, 18(11): 2433. |
| [32] | Li Y, Miao YY, Yuan HL, et al. Volatilome-based GWAS identifies OsWRKY19 and OsNAC021 as key regulators of rice aroma [J]. Mol Plant, 2024, 17(12): 1866-1882. |
| [1] | 樊荣辉, 罗远华, 陈艺荃, 方能炎, 陈燕, 钟淮钦, 叶秀仙. 文心兰‘金辉’和‘香水文心’花香形成比较分析[J]. 生物技术通报, 2026, 42(1): 105-113. |
| [2] | 刘语诗, 李镇, 邹宇琛, 汤维维, 李彬. 药用植物空间代谢组学研究进展[J]. 生物技术通报, 2025, 41(9): 22-31. |
| [3] | 刘建国, 刘格儿, 郭颖欣, 王斌, 王玉昆, 卢金凤, 黄文庭, 朱云娜. 转录组和代谢组联合解析‘桂柚1号’和‘沙田柚’果实品质差异[J]. 生物技术通报, 2025, 41(9): 168-181. |
| [4] | 刘泽洲, 段乃彬, 岳丽昕, 王清华, 姚行浩, 高莉敏, 孔素萍. 大蒜叶片蜡质成分分析及蜡质缺失基因Ggl-1筛选[J]. 生物技术通报, 2025, 41(9): 219-231. |
| [5] | 闫梦阳, 梁晓阳, 戴君昂, 张妍, 关团, 张辉, 刘良波, 孙志华. 阿莫西林降解菌的筛选及降解机制研究[J]. 生物技术通报, 2025, 41(9): 314-325. |
| [6] | 张雅祺, 王芹芹, 沈夏, 李旭苗, 高敏, 李军, 李辰, 王慧. 食管鳞状细胞癌早期进展风险的代谢物预警模型[J]. 生物技术通报, 2025, 41(9): 335-344. |
| [7] | 白雨果, 李婉迪, 梁建萍, 石志勇, 卢庚龙, 刘红军, 牛景萍. 哈茨木霉T9131对黄芪幼苗的促生机理[J]. 生物技术通报, 2025, 41(8): 175-185. |
| [8] | 柴军发, 洪波, 贾彦霞. 转录组和代谢组联合分析三株蜡蚧轮枝菌菌株毒力差异[J]. 生物技术通报, 2025, 41(8): 311-321. |
| [9] | 王月琛, 韩鑫骐, 魏文敏, 崔兆兰, 罗阳美, 陈鹏如, 王海岗, 刘龙龙, 张莉, 王纶. 黍稷落粒的生物学基础研究及落粒调控基因的鉴定[J]. 生物技术通报, 2025, 41(7): 164-171. |
| [10] | 蒋天威, 马培杰, 李亚娇, 陈才俊, 刘晓霞, 王小利. 二穗短柄草对光周期的代谢响应分析[J]. 生物技术通报, 2025, 41(7): 237-247. |
| [11] | 张越, 毕钰, 慕雪男, 郑子薇, 王志刚, 徐伟慧. 小麦赤霉病拮抗菌JB7的生防特性[J]. 生物技术通报, 2025, 41(7): 261-271. |
| [12] | 李成花, 豆飞飞, 任毓昭, 刘彩霞, 刘凤楼, 王掌军, 李清峰. 外施水杨酸对白粉菌侵染小麦的影响及白粉菌转录组分析[J]. 生物技术通报, 2025, 41(7): 272-280. |
| [13] | 郭秀娟, 冯宇, 吴瑞香, 王利琴, 杨建春. Ca2+处理对胡麻种子萌发影响的转录组分析[J]. 生物技术通报, 2025, 41(7): 139-149. |
| [14] | 黄旭升, 周雅莉, 柴旭东, 闻婧, 王计平, 贾小云, 李润植. 紫苏质体型PfLPAT1B基因的克隆及其在油脂合成中的功能分析[J]. 生物技术通报, 2025, 41(7): 226-236. |
| [15] | 李锐, 胡婷, 陈树溦, 王尧, 王计平. 紫苏PfMYB80转录因子正向调控花青素的生物合成[J]. 生物技术通报, 2025, 41(6): 243-255. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||