








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (5): 279-285.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0746
Previous Articles Next Articles
SUN Bao-zhen1( ), QUAN Long-ping1, KANG Hui1, YAO Yu-xin1, SHEN Tian2, CHEN Wei-ping2, DU Yuan-peng1, GAO Zhen1(
), QUAN Long-ping1, KANG Hui1, YAO Yu-xin1, SHEN Tian2, CHEN Wei-ping2, DU Yuan-peng1, GAO Zhen1( )
)
Received:2021-06-08
Online:2022-05-26
Published:2022-06-10
Contact:
GAO Zhen
E-mail:q864070339@163.com;gaoz89@sdau.edu.cn
SUN Bao-zhen, QUAN Long-ping, KANG Hui, YAO Yu-xin, SHEN Tian, CHEN Wei-ping, DU Yuan-peng, GAO Zhen. Back-splicing Primers-based PCR Method for Specific Detection of circRNA[J]. Biotechnology Bulletin, 2022, 38(5): 279-285.
| circRNA名称 Name of circRNA | 正向引物 Forward primer(5'-3') | 反向引物 Reverse primer(5'-3') |
|---|---|---|
| circRNA_4363 | AGAGATGCAGAACAACAGAGTTATG | GCGGGATTGAATCTGTGAATC |
| circRNA_6017 | CAGAAGGATACCGTGGAATCGAT | ACTCTGCTGCAGCCTGTAGCAT |
| circRNA_6044 | CATGCAACAGCTTTCAAGCAA | GAGCCTTCCTCCTTTCTTCCA |
| circRNA_7086 | TTTGCTGTTTCTGGATACGCTTCT | TGCAGCATCAACTCTTTCATTGTC |
Table 1 Primers for RT-PCR and qRT-PCR
| circRNA名称 Name of circRNA | 正向引物 Forward primer(5'-3') | 反向引物 Reverse primer(5'-3') |
|---|---|---|
| circRNA_4363 | AGAGATGCAGAACAACAGAGTTATG | GCGGGATTGAATCTGTGAATC |
| circRNA_6017 | CAGAAGGATACCGTGGAATCGAT | ACTCTGCTGCAGCCTGTAGCAT |
| circRNA_6044 | CATGCAACAGCTTTCAAGCAA | GAGCCTTCCTCCTTTCTTCCA |
| circRNA_7086 | TTTGCTGTTTCTGGATACGCTTCT | TGCAGCATCAACTCTTTCATTGTC |
| circRNA名称 Name of circRNA | 正向引物 Forward primer(5'-3') | 反向引物 Reverse primer(5'-3') |
|---|---|---|
| circRNA_4363 | ACTGATGGAGTTGATGAGGAA | GGTGTCCTTCTTCGGTTAAACT |
| circRNA_6017 | GCTCTGTAAACTTAACCAGGAAA | GCATGCCATCTGAGATAATCCT |
| circRNA_6044 | AAGAGAGAGCCAAGAAGGAGA | CCTTCTTGCTTGAAAGCTGT |
| circRNA_7086 | CGTTTTTTCTCAGAGAAGAAGG | ATGATGGAAGGATCAGTACCGT |
Table 2 Improved primers for RT-PCR and qRT-PCR
| circRNA名称 Name of circRNA | 正向引物 Forward primer(5'-3') | 反向引物 Reverse primer(5'-3') |
|---|---|---|
| circRNA_4363 | ACTGATGGAGTTGATGAGGAA | GGTGTCCTTCTTCGGTTAAACT |
| circRNA_6017 | GCTCTGTAAACTTAACCAGGAAA | GCATGCCATCTGAGATAATCCT |
| circRNA_6044 | AAGAGAGAGCCAAGAAGGAGA | CCTTCTTGCTTGAAAGCTGT |
| circRNA_7086 | CGTTTTTTCTCAGAGAAGAAGG | ATGATGGAAGGATCAGTACCGT |
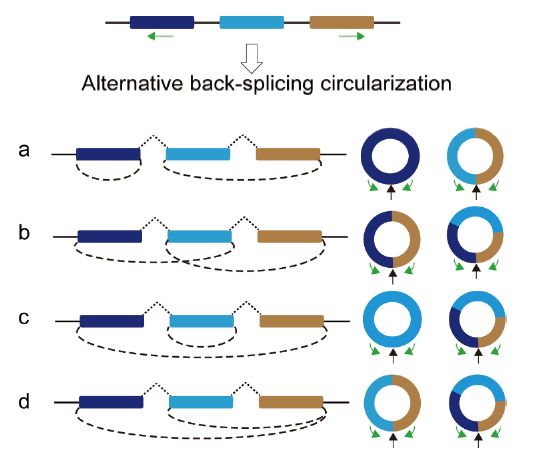
Fig. 1 Alternative back-splicing circularization of circRNA a : Juxtaposition relationship. b : Crossover relationship. c and d : Inclusion relatio-nship
| [1] |
Cocquerelle C, Mascrez B, Hétuin D, et al. Mis-splicing yields circular RNA molecules[J]. Faseb J, 1993, 7(1):155-160.
pmid: 7678559 |
| [2] | Chu QJ, Bai PP, Zhu XT, et al. Characteristics of plant circular RNAs[J]. Brief Bioinform, 2020, 21(1):135-143. |
| [3] |
Tan J, Zhou Z, Niu Y, et al. Identification and functional characterization of tomato CircRNAs derived from genes involved in fruit pigment accumulation[J]. Sci Rep, 2017, 7(1):8594.
doi: 10.1038/s41598-017-08806-0 URL |
| [4] |
Lu T, Cui L, Zhou Y, et al. Transcriptome-wide investigation of circular RNAs in rice[J]. RNA, 2015, 21(12):2076-2087.
doi: 10.1261/rna.052282.115 URL |
| [5] |
Zeng RF, Zhou JJ, Hu CG, et al. Transcriptome-wide identification and functional prediction of novel and flowering-related circular RNAs from trifoliate orange(Poncirus trifoliata L. Raf. )[J]. Planta, 2018, 247(5):1191-1202.
doi: 10.1007/s00425-018-2857-2 URL |
| [6] | Zheng WQ, Zhang Y, Chen B, et al. Identification and characterization of circRNAs in the developing stem cambium of poplar seedlings[J]. Mol Biol, 2020, 54(5):802-812. |
| [7] |
Ye CY, Zhang X, Chu Q, et al. Full-length sequence assembly reveals circular RNAs with diverse non-GT/AG splicing signals in rice[J]. RNA Biol, 2017, 14(8):1055-1063.
doi: 10.1080/15476286.2016.1245268 pmid: 27739910 |
| [8] |
Pan T, Sun X, Liu Y, et al. Heat stress alters genome-wide profiles of circular RNAs in Arabidopsis[J]. Plant Mol Biol, 2018, 96(3):217-229.
doi: 10.1007/s11103-017-0684-7 URL |
| [9] |
Zhao T, Wang L, Li S, et al. Characterization of conserved circular RNA in polyploid Gossypium species and their ancestors[J]. FEBS Lett, 2017, 591(21):3660-3669.
doi: 10.1002/1873-3468.12868 URL |
| [10] |
Tang B, Hao Z, Zhu Y, et al. Genome-wide identification and functional analysis of circRNAs in Zea mays[J]. PLoS One, 2018, 13(12):e0202375.
doi: 10.1371/journal.pone.0202375 URL |
| [11] | 高振. 葡萄circRNA鉴定分析及Vv-circATS1的抗冷功能研究[D]. 上海: 上海交通大学, 2019. |
| Gao Z. Identification and analysis of grape circRNAs and function research of Vv-circATS 1 on cold resistant[D]. Shanghai: Shanghai Jiaotong University, 2019 | |
| [12] |
Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency[J]. Nature, 2013, 495(7441):333-338.
doi: 10.1038/nature11928 URL |
| [13] |
Gao Y, Wang JF, Zhao FQ. CIRI:an efficient and unbiased algorithm for de novo circular RNA identification[J]. Genome Biol, 2015, 16(1):4.
doi: 10.1186/s13059-014-0571-3 URL |
| [14] |
Zhang XO, Wang HB, et al. Complementary sequence-mediated exon circularization[J]. Cell, 2014, 159(1):134-147.
doi: 10.1016/j.cell.2014.09.001 URL |
| [15] |
Gao Z, Li J, Luo M, et al. Characterization and cloning of grape circular RNAs identified the cold resistance-related Vv-circATS1[J]. Plant Physiol, 2019, 180(2):966-985.
doi: 10.1104/pp.18.01331 URL |
| [16] | Patop IL, Wüst S, Kadener S. Past, present, and future of circRNAs[J]. EMBO J, 2019, 38(16):e100836. |
| [1] | ZHAO Jie, LI An, LIANG Gang, JIN Xin-xin, PAN Li-gang. Research Progress in the Biological Functions of Plant circRNAs [J]. Biotechnology Bulletin, 2022, 38(10): 1-9. |
| [2] | FAN Ya-peng, RUI Cun, ZHANG Yue-xin, CHEN Xiu-gui, LU Xu-ke, WANG Shuai, ZHANG Hong, XU Nan, WANG Jing, CHEN Chao, YE Wu-wei. Cloning,Expression and Preliminary Bioinformatics Analysis of the Alkaline Tolerant Gene GhZAT12 in Gossypium hirsutum [J]. Biotechnology Bulletin, 2021, 37(8): 121-130. |
| [3] | CHEN Li-jie, YANG Fan, FAN Hai-yan, ZHAO Di, WANG Yuan-yuan, ZHU Xiao-feng, LIU Xiao-yu, DUAN Yu-xi. Advances of Non-coding RNA in Interactions Among Biocontrol Bacteria and Plant Nematodes and Host [J]. Biotechnology Bulletin, 2021, 37(7): 65-70. |
| [4] | PANG Peng-xiang, CHANG Yan-nan, YU Rui-min, GAO Gang. Cloning,Expression and Bioinformatics Analyses of StSRP1 Gene in Solanum tuberosum [J]. Biotechnology Bulletin, 2019, 35(7): 10-16. |
| [5] | ZHU Rui, YE Yu-qing, WANG Ya-xin, YANG Chen-ru, WANG Hong-wei, SUN Xiao-qing, ZHANG Yan, LI Shang-qi, LI Jiong-tang. Cloning,Expression and Comparative Analysis of Two Progesterone Receptor Genes in Cyprinus carpio [J]. Biotechnology Bulletin, 2019, 35(7): 46-53. |
| [6] | Hou Lan-fei, Yang Hong, Deng Zhi, Dai Long-jun, Men Zhong-hua, Li De-jun. Cloning,Expression and Bioinformatics Analyses of an ADC1 Gene in Hevea brasiliensis [J]. Biotechnology Bulletin, 2018, 34(11): 111-119. |
| [7] | YU Li-li, LIN Peng, GUO Song-lin, WANG Yi-lei, ZHANG Zi-ping, WANG Ting-ting, FENG Jian-jun. Molecular Cloning and Immune Function Analysis of TLR3 Gene in Anguilla japonica [J]. Biotechnology Bulletin, 2018, 34(1): 160-171. |
| [8] | XU Wen, YANG Hua-yu, ZHENG Yong-chang. Promoter-independent Transcription of Exogenous DNA and Subsequent RNA Splicing in Mammalian Cells [J]. Biotechnology Bulletin, 2017, 33(8): 139-145. |
| [9] | ZHANG Ying-yue ,MA Yue-hui ,ZHAO Qian-jun. Study Progress on Circular RNA [J]. Biotechnology Bulletin, 2017, 33(7): 29-34. |
| [10] | TANG Hui-rong, CUI Jin ,ZHANG Jia-hua, DAI You-guo ,LI Wei-ming ,LIAO Chen. Effects of PDSW Combined with Hyperthermia on COX-2 and Bcl-2 Expressions in Gastric Cancer Cells and Tissues [J]. Biotechnology Bulletin, 2017, 33(7): 224-230. |
| [11] | GUO Tian-lu, ZHANG Huan-huan, DU Jian-zhong, HAO Yao-shan, WANG Yi-xue, SUN Yi. mRNA Expression and Bioinformatics Analysis of Cysteine Synthase in Allium sativum [J]. Biotechnology Bulletin, 2016, 32(8): 96-102. |
| [12] | ZHANG Jia-jia, ZHANG Li-wei, LI Yong-peng, JIAO Xiao-lin, ZHU Feng-ling, DU Li. Evaluation of Candidate Reference Genes for Normalization of Quantitative RT-PCR in Cinnamomum camphora Under Various Abiotic Stresses [J]. Biotechnology Bulletin, 2016, 32(10): 205-211. |
| [13] | Yu Zhujun Chai Xiaojie Zhang Xiaolin Zhang Ting Xue Fei. Cloning and Expression Analysis of Small GTP-binding Protein Gene from Dunaliella salina(DsRab)Under Salt Stress [J]. Biotechnology Bulletin, 2013, 0(9): 77-83. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||