








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (3): 135-145.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0862
Previous Articles Next Articles
CHEN Xiao-song1( ), LIU Chao-jie1, ZHENG Jia2,3, QIAO Zong-wei2,3, LUO Hui-bo1, ZOU Wei1,2(
), LIU Chao-jie1, ZHENG Jia2,3, QIAO Zong-wei2,3, LUO Hui-bo1, ZOU Wei1,2( )
)
Received:2023-09-06
Online:2024-03-26
Published:2024-04-08
Contact:
ZOU Wei
E-mail:xschenyx@qq.com;weizou@suse.edu.cn
CHEN Xiao-song, LIU Chao-jie, ZHENG Jia, QIAO Zong-wei, LUO Hui-bo, ZOU Wei. Analyzing the Growth and Caproic Acid Metabolism Mechanism of Rummeliibacillus suwonensis 3B-1 by Tandem Mass Tag-based Quantitative Proteomics[J]. Biotechnology Bulletin, 2024, 40(3): 135-145.
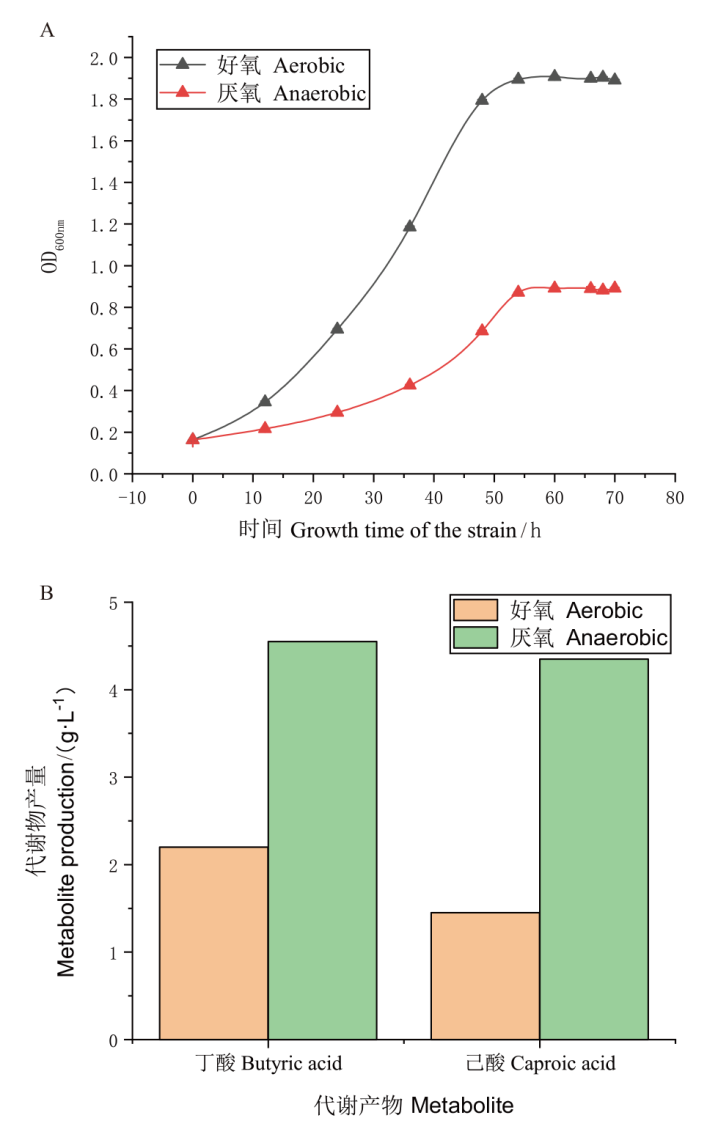
Fig. 1 Growth and metabolism of 3B-1 under aerobic and anaerobic conditions A : 3B-1 growth curves under aerobic and anaerobic conditions. B : Butyric acid and caproic acid production of 3B-1 under aerobic and anaerobic conditions
| 样品名称 Sample | 编号 Sample number | 蛋白浓度 Protein concentration/(μg·μL-1) | 蛋白总量 Total protein/μg |
|---|---|---|---|
| RH1 | 1 | 1.475 | 885 |
| RH2 | 2 | 3.197 | 1 918.2 |
| RH3 | 3 | 1.647 | 988.2 |
| RY1 | 4 | 1.268 | 760.8 |
| RY2 | 5 | 1.954 | 1 172.4 |
| RY3 | 6 | 1.608 | 964.8 |
Table 1 BCA protein quantification results
| 样品名称 Sample | 编号 Sample number | 蛋白浓度 Protein concentration/(μg·μL-1) | 蛋白总量 Total protein/μg |
|---|---|---|---|
| RH1 | 1 | 1.475 | 885 |
| RH2 | 2 | 3.197 | 1 918.2 |
| RH3 | 3 | 1.647 | 988.2 |
| RY1 | 4 | 1.268 | 760.8 |
| RY2 | 5 | 1.954 | 1 172.4 |
| RY3 | 6 | 1.608 | 964.8 |
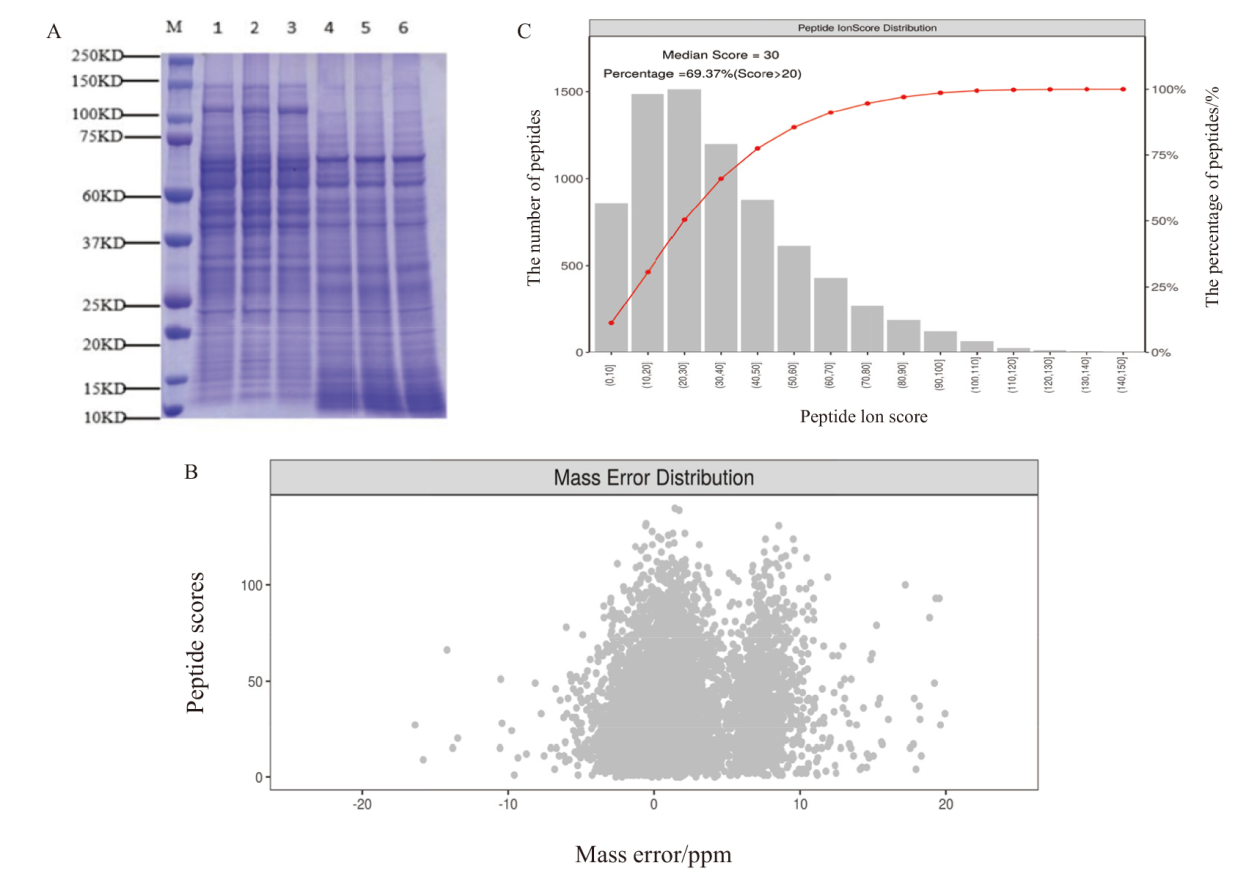
Fig. 2 Quality control of protein sample A: SDS-PAGE of protein samples. B: Distribution plot of peptide ion mass deviations. C: Distribution plot of peptide ion scores
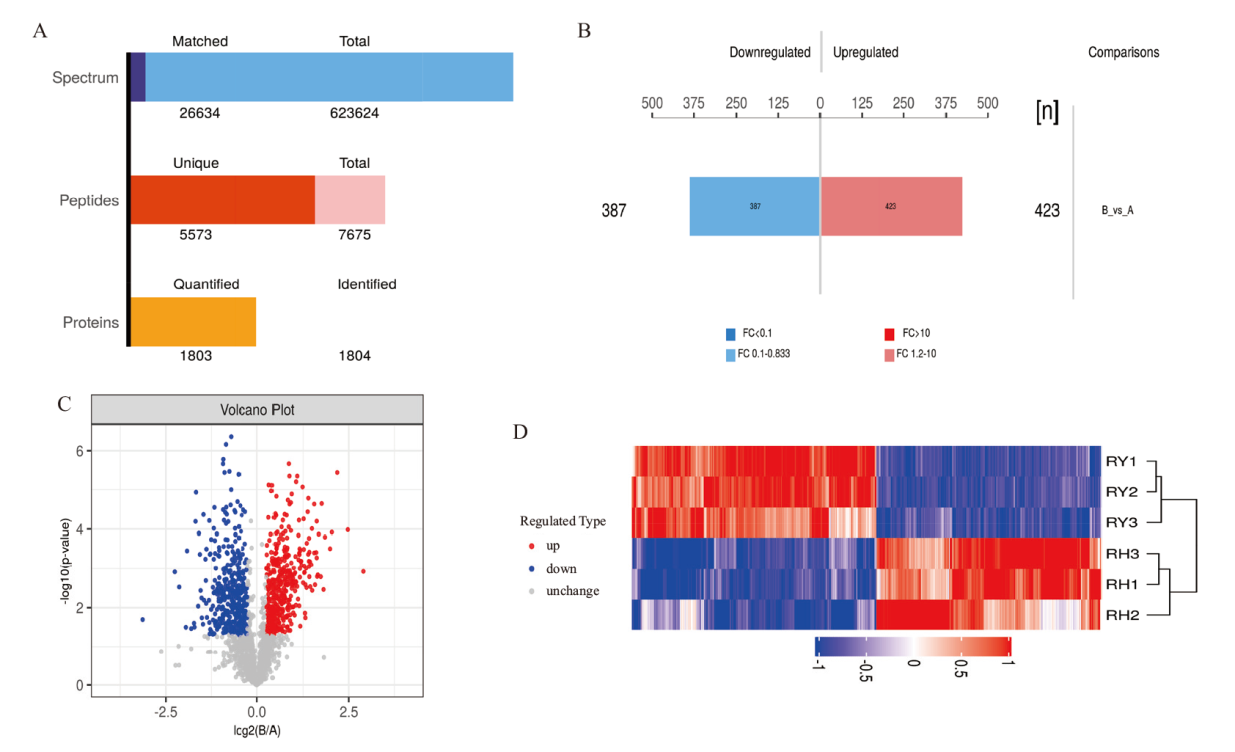
Fig. 3 Results of protein identification and differential expression A: Result graph of protein identification and quantification. B: Bar graph of protein quantification differences. C: Volcano plot of significant differences in the comparative group. D: Analysis graph of clustering of DEPs in the comparative group
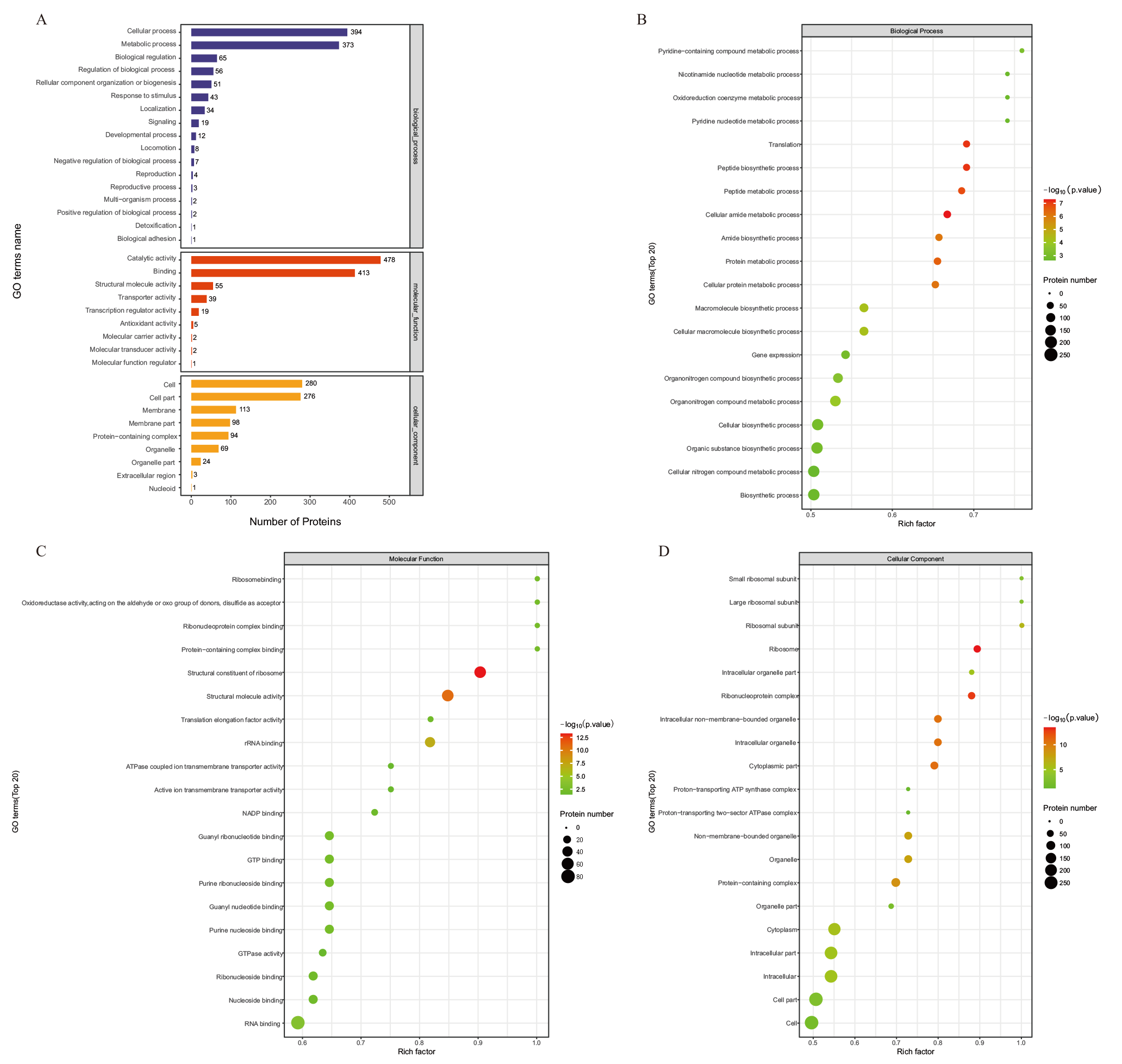
Fig. 4 GO annotation map of DEPs in comparative group A : GO annotation map of DEPs in the comparative group. B : GO functional enrichment bubble diagram under the biological process classification of DEPs in the comparative group. C: GO functional enrichment bubble diagram under the molecular function classification of DEPs in the comparative group. D : GO functional enrichment bubble diagram under the cell component classification of DEPs in the comparative group
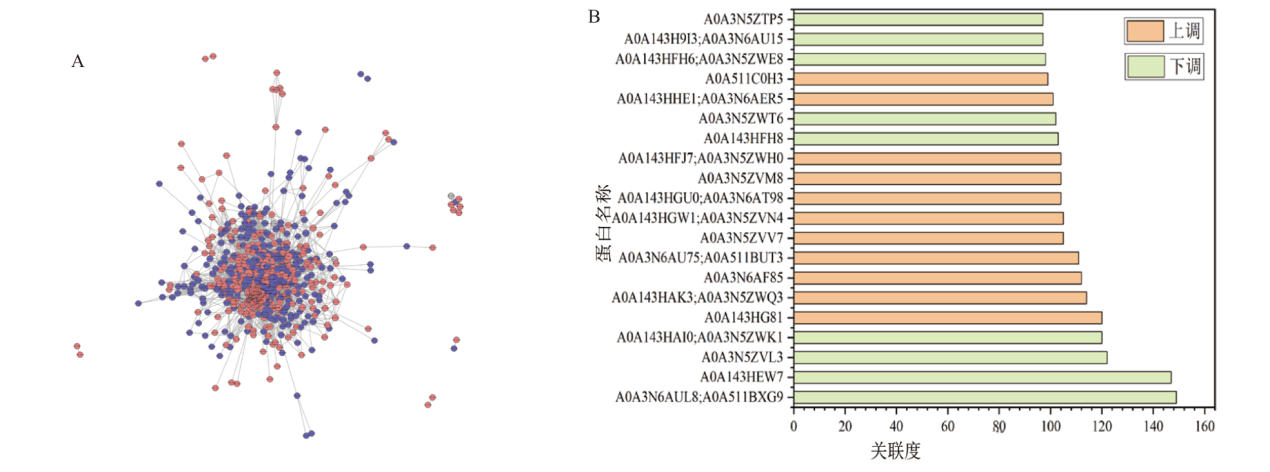
Fig. 6 Results of the interaction network analysis of DEP in the comparative groups A: Network interaction graph of DEPs in the comparative groups. B: Expression patterns of the top 20 proteins with the highest correlation in the DEPs interaction network
| 蛋白编号Protein | 基因名称Gene name | 蛋白描述Annotation |
|---|---|---|
| A0A3N6AUL8;A0A511BXG9 | pheT | Phenylalanine--tRNA ligase beta subunit |
| A0A143HEW7 | — | Riboflavin biosynthesis protein RibD |
| A0A3N5ZVL3 | rpoA | DNA-directed RNA polymerase subunit alpha |
| A0A143HAI0;A0A3N5ZWK1 | lepA | Elongation factor 4 |
| A0A143HG81 | tuf | Elongation factor Tu |
| A0A143HAK3;A0A3N5ZWQ3 | rpsB | 30S ribosomal protein S2 |
| A0A3N6AF85 | rplB | 50S ribosomal protein L2 |
| A0A3N6AU75;A0A511BUT3 | infB | Translation initiation factor IF-2 |
| A0A3N5ZVV7 | rplE | 50S ribosomal protein L5 |
| A0A143HGW1;A0A3N5ZVN4 | fusA | Elongation factor G |
| A0A143HGU0;A0A3N6AT98 | rpsE | 30S ribosomal protein S5 |
| A0A3N5ZVM8 | rpsC | 30S ribosomal protein S3 |
| A0A143HFJ7;A0A3N5ZWH0 | rpsG | 30S ribosomal protein S7 |
| A0A143HFH8 | rplD | 50S ribosomal protein L4 |
| A0A3N5ZWT6 | cinA | Putative competence-damage inducible |
| A0A143HHE1;A0A3N6AER5 | atpA | ATP synthase subunit alpha |
| A0A511C0H3 | rplR | 50S ribosomal protein L18 |
| A0A143HFH6;A0A3N5ZWE8 | rpsK | 30S ribosomal protein S11 |
| A0A143H9I3;A0A3N6AU15 | ftsY | Signal recognition particle receptor FtsY |
| A0A3N5ZTP5 | birA | Bifunctional ligase/repressor BirA |
Table 2 Top 20 proteins with the highest correlation in the interaction network of DEPs
| 蛋白编号Protein | 基因名称Gene name | 蛋白描述Annotation |
|---|---|---|
| A0A3N6AUL8;A0A511BXG9 | pheT | Phenylalanine--tRNA ligase beta subunit |
| A0A143HEW7 | — | Riboflavin biosynthesis protein RibD |
| A0A3N5ZVL3 | rpoA | DNA-directed RNA polymerase subunit alpha |
| A0A143HAI0;A0A3N5ZWK1 | lepA | Elongation factor 4 |
| A0A143HG81 | tuf | Elongation factor Tu |
| A0A143HAK3;A0A3N5ZWQ3 | rpsB | 30S ribosomal protein S2 |
| A0A3N6AF85 | rplB | 50S ribosomal protein L2 |
| A0A3N6AU75;A0A511BUT3 | infB | Translation initiation factor IF-2 |
| A0A3N5ZVV7 | rplE | 50S ribosomal protein L5 |
| A0A143HGW1;A0A3N5ZVN4 | fusA | Elongation factor G |
| A0A143HGU0;A0A3N6AT98 | rpsE | 30S ribosomal protein S5 |
| A0A3N5ZVM8 | rpsC | 30S ribosomal protein S3 |
| A0A143HFJ7;A0A3N5ZWH0 | rpsG | 30S ribosomal protein S7 |
| A0A143HFH8 | rplD | 50S ribosomal protein L4 |
| A0A3N5ZWT6 | cinA | Putative competence-damage inducible |
| A0A143HHE1;A0A3N6AER5 | atpA | ATP synthase subunit alpha |
| A0A511C0H3 | rplR | 50S ribosomal protein L18 |
| A0A143HFH6;A0A3N5ZWE8 | rpsK | 30S ribosomal protein S11 |
| A0A143H9I3;A0A3N6AU15 | ftsY | Signal recognition particle receptor FtsY |
| A0A3N5ZTP5 | birA | Bifunctional ligase/repressor BirA |
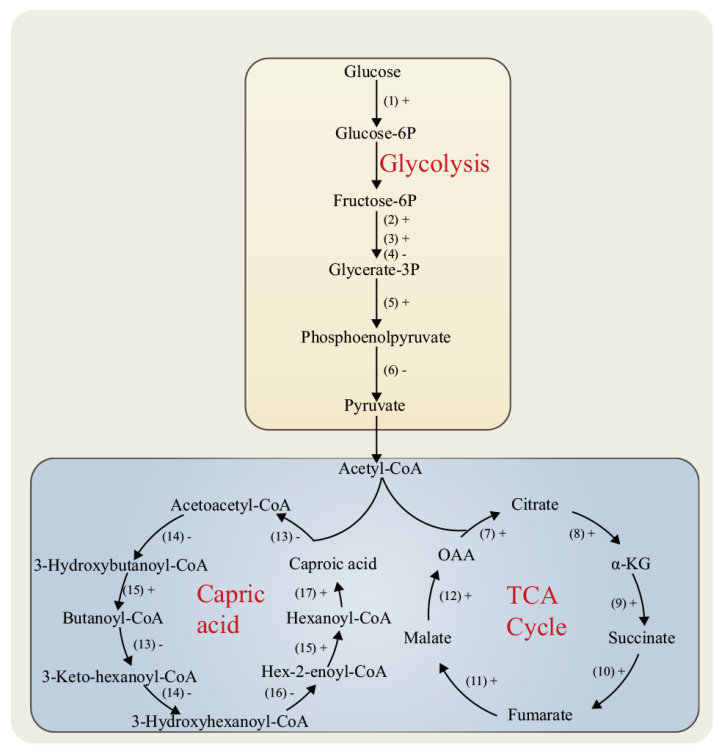
Fig. 7 Distribution of DEPs in the central metabolic and caproic acid metabolic pathways “ +” indicates that the expression of the enzyme is up-regulated, “-” indicates that the expression of the enzyme is down-regulated. (1): Hexokinase.(2): ATP-dependent 6-phosphofructokinase.(3): Phosphoglycerate kinase.(4): Fructose diphosphate aldolase.(5): 2-phosphoglycerate dehydratase.(6): Pyruvate kinase.(7): Citrate synthase.(8): Isocitrate dehydrogenase.(9): α-ketoglutarate dehydrogenase complex.(10): Succinate dehydrogenase.(11): Fumarase.(12): Malate dehydrogenase.(13): Acetyl-CoA acetyltransferase.(14): 3-hydroxybutyryl-CoA dehydrogenase.(15): Butyryl-CoA dehydrogenase.(16): 3-hydroxybutyryl-CoA dehydratase.(17): Acyl-CoA thioesterase
| [1] |
游玲, 简晓平, 范方勇, 等. 宜宾浓香型白酒产区窖泥生态监测[J]. 生物技术通报, 2023, 39(7): 254-265.
doi: 10.13560/j.cnki.biotech.bull.1985.2022-1406 |
|
You L, Jian XP, Fan FY, et al. Ecological monitoring of pit mud in Yibin strong-fragrance baijiu-producing region[J]. Biotechnol Bull, 2023, 39(7): 254-265.
doi: 10.13560/j.cnki.biotech.bull.1985.2022-1406 |
|
| [2] | 孙宝国, 吴继红, 黄明泉, 等. 白酒风味化学研究进展[J]. 中国食品学报, 2015, 15(9): 1-8. |
| Sun BG, Wu JH, Huang MQ, et al. Recent advances of flavor chemistry in Chinese liquor spirits(baijiu)[J]. J Chin Inst Food Sci Technol, 2015, 15(9): 1-8. | |
| [3] |
赵辉, 敞颜, 等. 浓香型白酒窖泥中高产己酸兼性厌氧细菌的分离鉴定[J]. 食品科学, 2012, 33(5): 177-182.
doi: 10.7506/spkx1002-6630-201205038 |
| Zhao H, Chang Y, et al. Isolation and identification of facultative anaerobic strains with high yield of hexanoic acid from luzhou-flavor liquor pit mud[J]. Food Sci, 2012, 33(5): 177-182. | |
| [4] |
Yao F, Yi B, Shen CH, et al. Chemical analysis of the Chinese liquor Luzhoulaojiao by comprehensive two-dimensional gas chromatography/time-of-flight mass spectrometry[J]. Sci Rep, 2015, 5: 9553.
doi: 10.1038/srep09553 pmid: 25857434 |
| [5] | 陈茂彬, 殷想想, 郭志豪, 等. 酪丁酸梭菌共培养对速生梭菌己酸代谢的影响及机理探讨[J]. 食品科学, 2023, 44(10): 158-164. |
|
Chen MB, Yin XX, et al. Effect and mechanism of co-culture with Clostridium tyrobutyricum on caproic acid mechanism in Clostridium celerecrescens[J]. Food Sci, 2023, 44(10): 158-164.
doi: 10.1111/jfds.1979.44.issue-1 URL |
|
| [6] |
Kenealy WR, Cao Y, Weimer PJ. Production of caproic acid by cocultures of ruminal cellulolytic bacteria and Clostridium kluyveri grown on cellulose and ethanol[J]. Appl Microbiol Biotechnol, 1995, 44(3-4): 507-513.
doi: 10.1007/s002530050590 pmid: 8597554 |
| [7] |
Dong WJ, Yang YL, Liu C, et al. Caproic acid production from anaerobic fermentation of organic waste - Pathways and microbial perspective[J]. Renew Sustain Energy Rev, 2023, 175: 113181.
doi: 10.1016/j.rser.2023.113181 URL |
| [8] | Debergh P, Van Dael M. Production of caproic acid from acetate and ethanol through microbial chain elongation: a techno-economic assessment[J]. Bioresour Technol Rep, 2022, 18: 101055. |
| [9] |
Agler MT, Spirito CM, Usack JG, et al. Chain elongation with reactor microbiomes: upgrading dilute ethanol to medium-chain carboxylates[J]. Energy Environ Sci, 2012, 5(8): 8189-8192.
doi: 10.1039/c2ee22101b URL |
| [10] |
Nzeteu CO, Coelho F, et al. Development of an enhanced chain elon-gation process for caproic acid production from waste-derived lactic acid and butyric acid[J]. J Clean Prod, 2022, 338: 130655.
doi: 10.1016/j.jclepro.2022.130655 URL |
| [11] |
Yang Q, Guo ST, Lu Q, et al. Butyryl/Caproyl-CoA: acetate CoA-transferase: cloning, expression and characterization of the key enzyme involved in medium-chain fatty acid biosynthesis[J]. Biosci Rep, 2021, 41(8): BSR20211135.
doi: 10.1042/BSR20211135 URL |
| [12] |
Her J, Kim J. Rummeliibacillus suwonensis sp. nov., isolated from soil collected in a mountain area of South Korea[J]. J Microbiol, 2013, 51(2): 268-272.
doi: 10.1007/s12275-013-3126-5 URL |
| [13] |
Hutchinson-Bunch C, Sanford JA, Hansen JR, et al. Assessment of TMT labeling efficiency in large-scale quantitative proteomics: the critical effect of sample pH[J]. ACS Omega, 2021, 6(19): 12660-12666.
doi: 10.1021/acsomega.1c00776 pmid: 34056417 |
| [14] |
Liu CJ, Du YF, Zheng J, et al. Production of caproic acid by Rummeliibacillus suwonensis 3B-1 isolated from the pit mud of strong-flavor Baijiu[J]. J Biotechnol, 2022, 358: 33-40.
doi: 10.1016/j.jbiotec.2022.08.017 URL |
| [15] |
Wiśniewski JR, Zougman A, Nagaraj N, et al. Universal sample preparation method for proteome analysis[J]. Nat Methods, 2009, 6(5): 359-362.
doi: 10.1038/nmeth.1322 pmid: 19377485 |
| [16] |
Sinclair J, Timms JF. Quantitative profiling of serum samples using TMT protein labelling, fractionation and LC-MS/MS[J]. Methods, 2011, 54(4): 361-369.
doi: 10.1016/j.ymeth.2011.03.004 pmid: 21397697 |
| [17] | 郭长明, 袁橙, 武彩红, 等. 应用iTRAQ定量蛋白质组学技术筛选无乳链球菌鱼源株与人源株差异表达蛋白[J]. 水产学报, 2018, 42(3): 442-451. |
| Guo CM, Yuan C, Wu CH, et al. Quantitative proteomic analysis of differential proteins in Streptococcus agalactiae piscine strain and human strain using iTRAQ[J]. J Fish China, 2018, 42(3): 442-451. | |
| [18] |
Rojas-Pirela M, Andrade-Alviárez D, et al. Phosphoglycerate kinase: structural aspects and functions, with special emphasis on the enzy-me from Kinetoplastea[J]. Open Biol, 2020, 10(11): 200302.
doi: 10.1098/rsob.200302 URL |
| [19] |
Yang LY, Wang ZX, Zhang AQ, et al. Reduction of the canonical function of a glycolytic enzyme enolase triggers immune responses that further affect metabolism and growth in Arabidopsis[J]. Plant Cell, 2022, 34(5): 1745-1767.
doi: 10.1093/plcell/koab283 URL |
| [20] |
Moxley WC, Eiteman MA. Pyruvate production by Escherichia coli by use of pyruvate dehydrogenase variants[J]. Appl Environ Microbiol, 2021, 87(13): e0048721.
doi: 10.1128/AEM.00487-21 URL |
| [21] |
Starling S. Pyruvate kinase regulates insulin secretion[J]. Nat Rev Endocrinol, 2021, 17(1): 3.
doi: 10.1038/s41574-020-00447-0 pmid: 33208920 |
| [22] |
Jacob H, Geng H, Shetty D, et al. Distinct interaction mechanism of RNA polymerase and ResD at proximal and distal subsites for transcription activation of nitrite reductase in Bacillus subtilis[J]. J Bacteriol, 2022, 204(2): e0043221.
doi: 10.1128/jb.00432-21 URL |
| [23] |
Nakano MM, Zuber P, Glaser P, et al. Two-component regulatory proteins ResD-ResE are required for transcriptional activation of fnr upon oxygen limitation in Bacillus subtilis[J]. J Bacteriol, 1996, 178(13): 3796-3802.
doi: 10.1128/jb.178.13.3796-3802.1996 pmid: 8682783 |
| [24] |
Michalska K, Jedrzejczak R, Wower J, et al. Mycobacterium tuberculosis Phe-tRNA synthetase: structural insights into tRNA recognition and aminoacylation[J]. Nucleic Acids Res, 2021, 49(9): 5351-5368.
doi: 10.1093/nar/gkab272 pmid: 33885823 |
| [25] |
Cong Q, Anishchenko I, Ovchinnikov S, et al. Protein interaction networks revealed by proteome coevolution[J]. Science, 2019, 365(6449): 185-189.
doi: 10.1126/science.aaw6718 pmid: 31296772 |
| [26] |
Sepúlveda-Cisternas I, Lozano Aguirre L, Fuentes Flores A, et al. Transcriptomics reveals a cross-modulatory effect between riboflavin and iron and outlines responses to riboflavin biosynthesis and uptake in Vibrio cholerae[J]. Sci Rep, 2018, 8(1): 3149.
doi: 10.1038/s41598-018-21302-3 pmid: 29453341 |
| [27] |
Yu S, Meng SM, Xiang MX, et al. Phosphoenolpyruvate carboxykinase in cell metabolism: roles and mechanisms beyond gluconeogenesis[J]. Mol Metab, 2021, 53: 101257.
doi: 10.1016/j.molmet.2021.101257 URL |
| [28] |
Britt EC, Lika J, Giese MA, et al. Switching to the cyclic pentose phosphate pathway powers the oxidative burst in activated neutrophils[J]. Nat Metab, 2022, 4(3): 389-403.
doi: 10.1038/s42255-022-00550-8 pmid: 35347316 |
| [29] |
Carvajal-Arroyo JM, Candry P, Andersen SJ, et al. Granular fermentation enables high rate caproic acid production from solid-free thin stillage[J]. Green Chem, 2019, 21(6): 1330-1339.
doi: 10.1039/c8gc03648a |
| [30] |
Chwialkowska J, Duber A, Zagrodnik R, et al. Caproic acid production from acid whey via open culture fermentation - Evaluation of the role of electron donors and downstream processing[J]. Bioresour Technol, 2019, 279: 74-83.
doi: 10.1016/j.biortech.2019.01.086 URL |
| [31] |
徐友强, 孙宝国, 蒋玥凤, 等. 基于比较基因组学的Clostrid-ium kluyveri己酸代谢途径关键酶生物信息学分析[J]. 食品科学, 2019, 40(4): 122-129.
doi: 10.7506/spkx1002-6630-20180413-183 |
| Xu YQ, Sun BG, Jiang YF, et al. Bioinformatics analysis of the key enzymes of the hexanoic acid metabolic pathway in Clostridium kluyveri based on comparative genomics[J]. Food Sci, 2019, 40(4): 122-129. |
| [1] | YANG Wei-jie, YANG Zhou-lin, ZHU Hao-dong, WEI Yu, LIU Jun, LIU Xun. Study on the Properties and Functions of LchAD Protein, a Key Module of Lichenysin Synthase [J]. Biotechnology Bulletin, 2024, 40(3): 322-332. |
| [2] | GONG Li-li, YU Hua, YANG Jie, CHEN Tian-chi, ZHAO Shuang-ying, WU Yue-yan. Identification and Analysis of Grape(Vitis vinifera L.)CYP707A Gene Family and Functional Verification to Fruit Ripening [J]. Biotechnology Bulletin, 2024, 40(2): 160-171. |
| [3] | ZHANG Lu-yang, HAN Wen-long, XU Xiao-wen, YAO Jian, LI Fang-fang, TIAN Xiao-yuan, ZHANG Zhi-qiang. Identification and Expression Analysis of the Tobacco TCP Gene Family [J]. Biotechnology Bulletin, 2023, 39(6): 248-258. |
| [4] | LI Jing-rui, WANG Yu-bo, XIE Zi-wei, LI Chang, WU Xiao-lei, GONG Bin-bin, GAO Hong-bo. Identification and Expression Analysis of PIN Gene Family in Melon Under High Temperature Stress [J]. Biotechnology Bulletin, 2023, 39(5): 192-204. |
| [5] | GUO San-bao, SONG Mei-ling, LI Ling-xin, YAO Zi-zhao, GUI Ming-ming, HUANG Sheng-he. Cloning and Analysis of Chalcone Synthase Gene and Its Promoter from Euphorbia maculata [J]. Biotechnology Bulletin, 2023, 39(4): 148-156. |
| [6] | WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(4): 246-258. |
| [7] | YANG Lan, ZHANG Chen-xi, FAN Xue-wei, WANG Yang-guang, WANG Chun-xiu, LI Wen-ting. Gene Cloning, Expression Pattern, and Promoter Activity Analysis of Chicken BMP15 [J]. Biotechnology Bulletin, 2023, 39(4): 304-312. |
| [8] | CHEN Qiang, ZHOU Ming-kang, SONG Jia-min, ZHANG Chong, WU Long-kun. Identification and Analysis of LBD Gene Family and Expression Analysis of Fruit Development in Cucumis melo [J]. Biotechnology Bulletin, 2023, 39(3): 176-183. |
| [9] | PING Huai-lei, GUO Xue, YU Xiao, SONG Jing, DU Chun, WANG Juan, ZHANG Huai-bi. Cloning and Expression of PdANS in Paeonia delavayi and Correlation with Anthocyanin Content [J]. Biotechnology Bulletin, 2023, 39(3): 206-217. |
| [10] | XING Yuan, SONG Jian, LI Jun-yi, ZHENG Ting-ting, LIU Si-chen, QIAO Zhi-jun. Identification of AP Gene Family and Its Response Analysis to Abiotic Stress in Setaria italica [J]. Biotechnology Bulletin, 2023, 39(11): 238-251. |
| [11] | CHEN Chu-yi, YANG Xiao-mei, CHEN Sheng-yan, CHEN Bin, YUE Li-ran. Expression Analysis of the ZF-HD Gene Family in Chrysanthemum nankingense Under Drought and ABA Treatment [J]. Biotechnology Bulletin, 2023, 39(11): 270-282. |
| [12] | YANG Min, LONG Yu-qing, ZENG Juan, ZENG Mei, ZHOU Xin-ru, WANG Ling, FU Xue-sen, ZHOU Ri-bao, LIU Xiang-dan. Cloning and Function Analysis of Gene UGTPg17 and UGTPg36 in Lonicera macranthoides [J]. Biotechnology Bulletin, 2023, 39(10): 256-267. |
| [13] | GUO Zhi-hao, JIN Ze-xin, LIU Qi, GAO Li. Bioinformatics Analysis, Subcellular Localization and Toxicity Verification of Effector g11335 in Tilletia contraversa Kühn [J]. Biotechnology Bulletin, 2022, 38(8): 110-117. |
| [14] | YU Qiu-lin, MA Jing-yi, ZHAO Pan, SUN Peng-fang, HE Yu-mei, LIU Shi-biao, GUO Hui-hong. Cloning and Functional Analysis of Gynostemma pentaphyllum GpMIR156a and GpMIR166b [J]. Biotechnology Bulletin, 2022, 38(7): 186-193. |
| [15] | CHEN Jia-min, LIU Yong-jie, MA Jin-xiu, LI Dan, GONG Jie, ZHAO Chang-ping, GENG Hong-wei, GAO Shi-qing. Expression Pattern Analysis of Histone Methyltransferase Under Drought Stress in Hybrid Wheat [J]. Biotechnology Bulletin, 2022, 38(7): 51-61. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||