








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (7): 1-18.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0193
LIU Wen-hao1( ), WU Liu-ji2, XU Fang1,3(
), WU Liu-ji2, XU Fang1,3( )
)
Received:2024-02-29
Online:2024-07-26
Published:2024-07-30
Contact:
XU Fang
E-mail:liuwenhao2k99@163.com;fxu@sdu.edu.cn
LIU Wen-hao, WU Liu-ji, XU Fang. Regulatory Mechanisms of Small Peptides in Plant Meristem Development and Its Research Advances in Crop Improvement[J]. Biotechnology Bulletin, 2024, 40(7): 1-18.
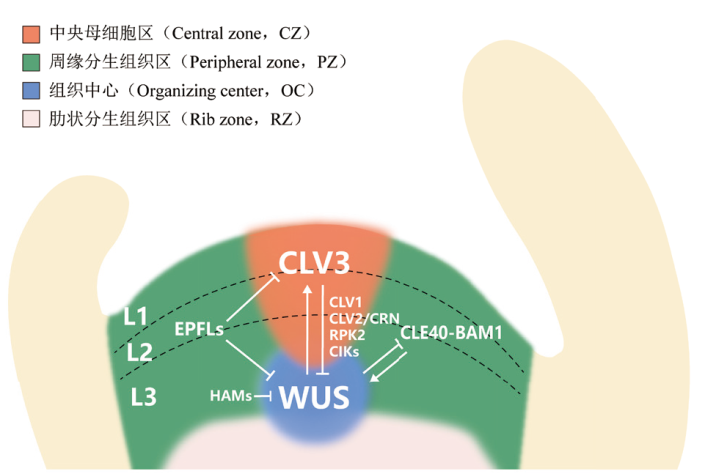
Fig. 1 Molecular mechanisms involved in the regulation of shoot apical meristem(SAM)by small peptides in Arabidopsis In Arabidopsis, the shoot apical meristem(SAM)is composed of organizing center(OC), central zone(CZ), peripheral zone(PZ), and rib zone(RZ), which can also be divided into layer L1, L2, and L3. The key transcription factor WUS, which maintains stem cell characteristics in SAM, is specifically expressed in OC and migrates to the CZ through plasmodesmata. WUS promotes the expression of CLV3 in the CZ under the restriction of the transcription factor HAM. After being modified and cleaved, mature CLV3 peptides move to the OC and inhibits the expression of WUS, establishing a key negative feedback pathway in SAM regulation. CLV1, CLV2, CRN, RPK2, and CIKs form different receptor complexes that recognize CLV3 peptide and transmit signal to inhibit WUS expression. CLE40 and its receptor BAM1 are expressed in the PZ, forming an additional negative feedback pathway with WUS. EPFL and its receptor ERf are expressed in the PZ, restricting the expression of WUS and CLV3 to the CZ
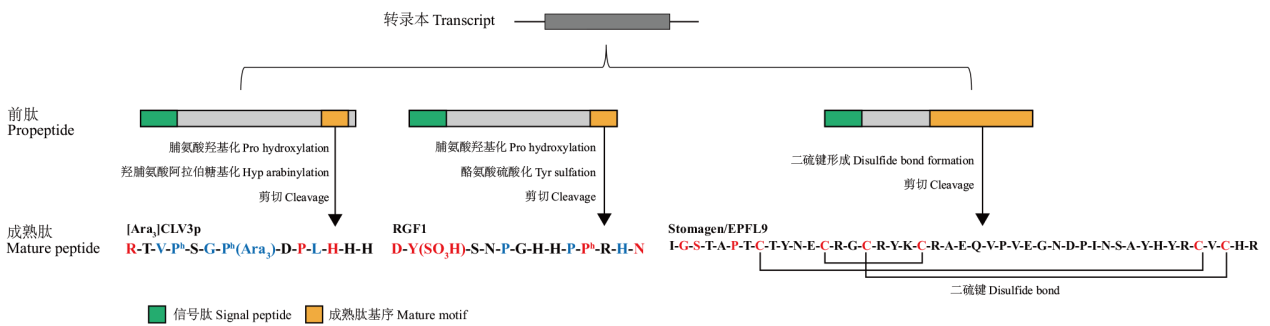
Fig. 2 Post-translational modifications and sequence characteristics of R-type CLE, RGF, EPF/EPFL peptides CLV3, RGF1 and Stomagen/EPFL9 were used as examples to demonstrate the post-translational modifications and sequence characteristics of R-type CLE, RGF, EPF/EPFL peptides[16,63 -64]. Identical amino acid residues are highlighted in red, and similar amino acid residues are highlighted in blue. The first amino acid of the mature peptides of H-type CLE is His, including TDIF(CLE41/44/42)and CLE46. The other CLE peptides are R-type CLE peptides, and the first amino acid of the mature peptides R-type is Arg[16,65]
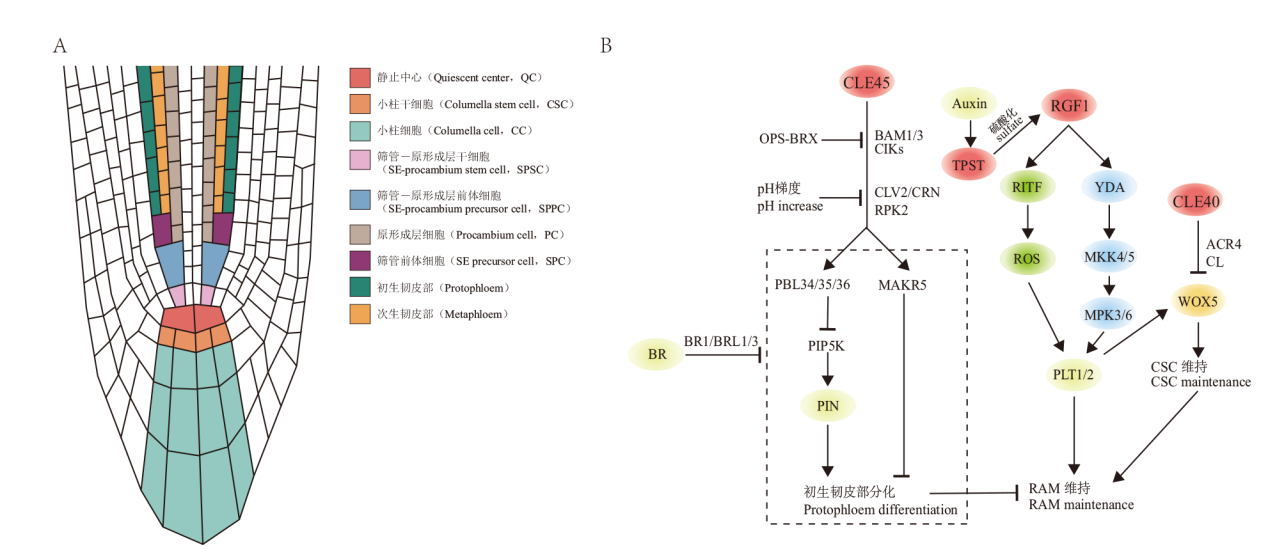
Fig. 3 Molecular mechanisms involved in the regulation of root apical meristem(RAM)by small peptides in Arabidopsis A:In Arabidopsis, the root apical meristem(RAM)comprises of the quiescent center(QC), distal meristem(DM), and proximal meristem(PM). The DM consists of columella stem cells(CSCs)and the PM includes a layer of stem cells adjacent to QC and its progeny cells. Protophloem in the PM is produced by Sieve Element(SE)-Procambium Stem Cell(SPSC)after multiple periclinal and anticlinal divisions. B:CLE40-CLV1/ACR4 inhibits WUS to promote the differentiation of CSCs, and CLE45 inhibits the differentiation of the protophloem through different receptor complexes composed of BAM1/3, CIKs, CLV2/CRN, and RPK2, with downstream components including MAKR5 and PBL34/35/36. OPS, BRX, BR signals, and pH gradient are all involved in the CLE45 signaling pathway, regulating protophloem differentiation. RGF1-RGFR upregulates PLT1/2 through MAPK cascades and ROS to maintain meristem proliferation activity in RAM. The tyrosine sulfation of RGF1 is catalyzed by TPST, and auxin signals indirectly regulate the RGF1 signaling pathway by upregulating TPST
| 定位 Location | 家族 Family | 物种 Species | 小肽 Peptide | 受体 Receptor | 下游组分 Downstream components | 功能 Functions | 参考文献 References |
|---|---|---|---|---|---|---|---|
| SAM | CLE | 拟南芥 | CLV3 | CLV1,CLV2,CRN,RPK2,CIKs | PBL34/35/36,MAPK级联,Ca2+,KAPP,POL/PLL,AGB1,WUS | 抑制WUS的表达,抑制茎尖、花序、花分生组织干细胞活性,调控器官产生 | [ |
| CLE40 | BAM1 | WUS | 作为来自周缘分生组织区的信号,微调WUS的表达和干细胞活性 | [ | |||
| 水稻 | FON2 (FON4) | FON1 | 抑制花分生组织干细胞活性,调控枝梗、花器官和雌蕊数量等产量 性状 | [ | |||
| FOS1 | 与FON2-FON1冗余性地调控花分生组织干细胞活性 | [ | |||||
| FCP1/2 | OsWOX4 | 抑制OsWOX4的表达,抑制茎尖分生组织干细胞活性 | [ | ||||
| 玉米 | ZmFCP1 | FEA2,FEA3 | ZmCRN | 抑制茎尖、花序分生组织干细胞活性,调控玉米果穗大小、穗行数、行粒数等产量性状 | [ | ||
| ZmCLE7 | FEA2 | CT2 | 抑制茎尖、花序分生组织干细胞活性,调控玉米果穗大小、穗行数、行粒数等产量性状 | [ | |||
| ZmCLE1E5 | 与ZmCLE7冗余性地调控花序分生组织干细胞活性,调控玉米果穗大小、穗行数、行粒数等产量性状 | [ | |||||
| 番茄 | SlCLV3 | FAB | SLWUS | 抑制SlWUS的表达,抑制茎尖、花序、花分生组织干细胞活性,调控花、花心皮数量,影响果实大小 | [ | ||
| SlCLE9 | FAB | SLWUS | 与SlCLV3冗余性地调控分生组织干细胞活性,调控花、花心皮数量,影响果实大小 | [ | |||
| EPF/ EPFL | 拟南芥 | EPFL1/2/4/6 | ERf | WUS,CLV3 | 限制WUS和CLV3的表达区域,调控茎尖分生组织稳态 | [ | |
| 水稻 | GAD1(OsEPFL1) | 调控水稻每穗粒数、粒形和芒的发育 | [ | ||||
| OsEPFL6/7/8/9 | OsER1 | MAPK级联 | 调节水稻每穗粒数 | [ | |||
| RAM | CLE | 拟南芥 | CLE40/16/ 17 | ACR4,CLV1,CIKs | WOX5 | 抑制WOX5表达,促进小柱干细胞的分化,调控根远端分生组织稳态 | [ |
| CLE25/26/33/45 | BAM1/3,CLV2,CRN,CIKs | PBL34/35/36,BRX,OPS | 抑制初生韧皮部分化,调控根近端分生组织稳态 | [ | |||
| CLE14 | CLV2,PEPR2 | SCR,SHR,PIN | 通过调控根分生组织活性参与低磷胁迫响应 | [ | |||
| RGF | 拟南芥 | RGF1/2/3 | RGFR1/2/3/4/5 | RITF1,MAPK级联,PLT1/2,WOX5 | 响应生长素信号,通过下游PLT1/PLT2和WOX5调控根近端和远端分生组织稳态,并参与低磷胁迫 响应 | [ |
Table 1 Function of small peptides in apical meristem regulation
| 定位 Location | 家族 Family | 物种 Species | 小肽 Peptide | 受体 Receptor | 下游组分 Downstream components | 功能 Functions | 参考文献 References |
|---|---|---|---|---|---|---|---|
| SAM | CLE | 拟南芥 | CLV3 | CLV1,CLV2,CRN,RPK2,CIKs | PBL34/35/36,MAPK级联,Ca2+,KAPP,POL/PLL,AGB1,WUS | 抑制WUS的表达,抑制茎尖、花序、花分生组织干细胞活性,调控器官产生 | [ |
| CLE40 | BAM1 | WUS | 作为来自周缘分生组织区的信号,微调WUS的表达和干细胞活性 | [ | |||
| 水稻 | FON2 (FON4) | FON1 | 抑制花分生组织干细胞活性,调控枝梗、花器官和雌蕊数量等产量 性状 | [ | |||
| FOS1 | 与FON2-FON1冗余性地调控花分生组织干细胞活性 | [ | |||||
| FCP1/2 | OsWOX4 | 抑制OsWOX4的表达,抑制茎尖分生组织干细胞活性 | [ | ||||
| 玉米 | ZmFCP1 | FEA2,FEA3 | ZmCRN | 抑制茎尖、花序分生组织干细胞活性,调控玉米果穗大小、穗行数、行粒数等产量性状 | [ | ||
| ZmCLE7 | FEA2 | CT2 | 抑制茎尖、花序分生组织干细胞活性,调控玉米果穗大小、穗行数、行粒数等产量性状 | [ | |||
| ZmCLE1E5 | 与ZmCLE7冗余性地调控花序分生组织干细胞活性,调控玉米果穗大小、穗行数、行粒数等产量性状 | [ | |||||
| 番茄 | SlCLV3 | FAB | SLWUS | 抑制SlWUS的表达,抑制茎尖、花序、花分生组织干细胞活性,调控花、花心皮数量,影响果实大小 | [ | ||
| SlCLE9 | FAB | SLWUS | 与SlCLV3冗余性地调控分生组织干细胞活性,调控花、花心皮数量,影响果实大小 | [ | |||
| EPF/ EPFL | 拟南芥 | EPFL1/2/4/6 | ERf | WUS,CLV3 | 限制WUS和CLV3的表达区域,调控茎尖分生组织稳态 | [ | |
| 水稻 | GAD1(OsEPFL1) | 调控水稻每穗粒数、粒形和芒的发育 | [ | ||||
| OsEPFL6/7/8/9 | OsER1 | MAPK级联 | 调节水稻每穗粒数 | [ | |||
| RAM | CLE | 拟南芥 | CLE40/16/ 17 | ACR4,CLV1,CIKs | WOX5 | 抑制WOX5表达,促进小柱干细胞的分化,调控根远端分生组织稳态 | [ |
| CLE25/26/33/45 | BAM1/3,CLV2,CRN,CIKs | PBL34/35/36,BRX,OPS | 抑制初生韧皮部分化,调控根近端分生组织稳态 | [ | |||
| CLE14 | CLV2,PEPR2 | SCR,SHR,PIN | 通过调控根分生组织活性参与低磷胁迫响应 | [ | |||
| RGF | 拟南芥 | RGF1/2/3 | RGFR1/2/3/4/5 | RITF1,MAPK级联,PLT1/2,WOX5 | 响应生长素信号,通过下游PLT1/PLT2和WOX5调控根近端和远端分生组织稳态,并参与低磷胁迫 响应 | [ |
| [1] | Heidstra R, Sabatini S. Plant and animal stem cells: similar yet different[J]. Nat Rev Mol Cell Biol, 2014, 15(5): 301-312. |
| [2] |
Wang Y, Jiao YL. Cell signaling in the shoot apical meristem[J]. Plant Physiol, 2023, 193(1): 70-82.
doi: 10.1093/plphys/kiad309 pmid: 37224874 |
| [3] |
Motte H, Vanneste S, Beeckman T. Molecular and environmental regulation of root development[J]. Annu Rev Plant Biol, 2019, 70: 465-488.
doi: 10.1146/annurev-arplant-050718-100423 pmid: 30822115 |
| [4] | 蔺欢, 王俊娟, 孙振婷, 等. 植物小分子肽的研究进展[J]. 西北植物学报, 2021, 41(1): 168-180. |
| Lin H, Wang JJ, Sun ZT, et al. The research progress of plant small molecular peptides[J]. Acta Bot Boreali Occidentalia Sin, 2021, 41(1): 168-180. | |
| [5] | Song XF, Hou XL, Liu CM. CLE peptides: critical regulators for stem cell maintenance in plants[J]. Planta, 2021, 255(1): 5. |
| [6] |
Lindsay P, Swentowsky KW, Jackson D. Cultivating potential: Harnessing plant stem cells for agricultural crop improvement[J]. Mol Plant, 2024, 17(1): 50-74.
doi: 10.1016/j.molp.2023.12.014 pmid: 38130059 |
| [7] |
Perales M, Reddy GV. Stem cell maintenance in shoot apical meristems[J]. Curr Opin Plant Biol, 2012, 15(1): 10-16.
doi: 10.1016/j.pbi.2011.10.008 pmid: 22079787 |
| [8] |
Xie MT, Tataw M, Venugopala Reddy G. Towards a functional understanding of cell growth dynamics in shoot meristem stem-cell niche[J]. Semin Cell Dev Biol, 2009, 20(9): 1126-1133.
doi: 10.1016/j.semcdb.2009.09.014 pmid: 19782146 |
| [9] | 陆维超, 赵建国, 张莉, 等. 植物茎尖分生组织分化调控机制研究进展[J]. 西北植物学报, 2016, 36(5): 1055-1065. |
| Lu WC, Zhao JG, Zhang L, et al. Differentiation and regulation of the shoot apical meristem[J]. Acta Bot Boreali Occidentalia Sin, 2016, 36(5): 1055-1065. | |
| [10] |
Cock JM, McCormick S. A large family of genes that share homology with CLAVATA3[J]. Plant Physiol, 2001, 126(3): 939-942.
doi: 10.1104/pp.126.3.939 pmid: 11457943 |
| [11] | Goad DM, Zhu CM, Kellogg EA. Comprehensive identification and clustering of CLV3/ESR-related(CLE)genes in plants finds groups with potentially shared function[J]. New Phytol, 2017, 216(2): 605-616. |
| [12] |
Carbonnel S, Cornelis S, Hazak O. The CLE33 peptide represses phloem differentiation via autocrine and paracrine signaling in Arabidopsis[J]. Commun Biol, 2023, 6(1): 588.
doi: 10.1038/s42003-023-04972-2 pmid: 37280369 |
| [13] |
Sharma VK, Ramirez J, Fletcher JC. The Arabidopsis CLV3-like(CLE)genes are expressed in diverse tissues and encode secreted proteins[J]. Plant Mol Biol, 2003, 51(3): 415-425.
doi: 10.1023/a:1022038932376 pmid: 12602871 |
| [14] | Jun J, Fiume E, Roeder AHK, et al. Comprehensive analysis of CLE polypeptide signaling gene expression and overexpression activity in Arabidopsis[J]. Plant Physiol, 2010, 154(4): 1721-1736. |
| [15] | 吕倩雯, 杨永芳. 植物小肽信号生物学功能及其在作物改良中研究进展[J]. 遗传, 2023, 45(9): 813-828. |
| Lv QW, Yang YF. The biological functions of peptide signaling in plant and the advances on its utilization for crop improvement[J]. Hereditas: Beijing, 2023, 45(9): 813-828. | |
| [16] |
Ito Y, Nakanomyo I, Motose H, et al. Dodeca-CLE peptides as suppressors of plant stem cell differentiation[J]. Science, 2006, 313(5788): 842-845.
doi: 10.1126/science.1128436 pmid: 16902140 |
| [17] |
Kinoshita A, Nakamura Y, Sasaki E, et al. Gain-of-function phenotypes of chemically synthetic CLAVATA3/ESR-related(CLE)peptides in Arabidopsis thaliana and Oryza sativa[J]. Plant Cell Physiol, 2007, 48(12): 1821-1825.
doi: 10.1093/pcp/pcm154 pmid: 17991631 |
| [18] |
Laux T, Mayer KF, Berger J, et al. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis[J]. Development, 1996, 122(1): 87-96.
doi: 10.1242/dev.122.1.87 pmid: 8565856 |
| [19] |
Mayer KF, Schoof H, Haecker A, et al. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem[J]. Cell, 1998, 95(6): 805-815.
doi: 10.1016/s0092-8674(00)81703-1 pmid: 9865698 |
| [20] | Yadav RK, Perales M, Gruel J, et al. WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex[J]. Genes Dev, 2011, 25(19): 2025-2030. |
| [21] |
Zhou Y, Yan A, Han H, et al. HAIRY MERISTEM with WUSCHEL confines CLAVATA3 expression to the outer apical meristem layers[J]. Science, 2018, 361(6401): 502-506.
doi: 10.1126/science.aar8638 pmid: 30072538 |
| [22] |
Schoof H, Lenhard M, Haecker A, et al. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes[J]. Cell, 2000, 100(6): 635-644.
doi: 10.1016/s0092-8674(00)80700-x pmid: 10761929 |
| [23] |
Brand U, Fletcher JC, Hobe M, et al. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity[J]. Science, 2000, 289(5479): 617-619.
doi: 10.1126/science.289.5479.617 pmid: 10915624 |
| [24] | Clark SE, Running MP, Meyerowitz EM. CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1[J]. Development, 1995, 121(7): 2057-2067. |
| [25] |
Fletcher JC, Brand U, Running MP, et al. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems[J]. Science, 1999, 283(5409): 1911-1914.
doi: 10.1126/science.283.5409.1911 pmid: 10082464 |
| [26] | Schlegel J, Denay G, Wink R, et al. Control of Arabidopsis shoot stem cell homeostasis by two antagonistic CLE peptide signalling pathways[J]. eLife, 2021, 10: e70934. |
| [27] | Hobe M, Müller R, Grünewald M, et al. Loss of CLE40, a protein functionally equivalent to the stem cell restricting signal CLV3, enhances root waving in Arabidopsis[J]. Dev Genes Evol, 2003, 213(8): 371-381. |
| [28] |
Ni J, Clark SE. Evidence for functional conservation, sufficiency, and proteolytic processing of the CLAVATA3 CLE domain[J]. Plant Physiol, 2006, 140(2): 726-733.
doi: 10.1104/pp.105.072678 pmid: 16407446 |
| [29] |
Ogawa M, Shinohara H, Sakagami Y, et al. Arabidopsis CLV3 peptide directly binds CLV1 ectodomain[J]. Science, 2008, 319(5861): 294.
doi: 10.1126/science.1150083 pmid: 18202283 |
| [30] | Fiers M, Golemiec E, Xu J, et al. The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway[J]. Plant Cell, 2005, 17(9): 2542-2553. |
| [31] |
Yamaguchi YL, Ishida T, Yoshimura M, et al. A collection of mutants for CLE-peptide-encoding genes in Arabidopsis generated by CRISPR/Cas9-mediated gene targeting[J]. Plant Cell Physiol, 2017, 58(11): 1848-1856.
doi: 10.1093/pcp/pcx139 pmid: 29036337 |
| [32] |
Rodriguez-Leal D, Xu C, Kwon CT, et al. Evolution of buffering in a genetic circuit controlling plant stem cell proliferation[J]. Nat Genet, 2019, 51(5): 786-792.
doi: 10.1038/s41588-019-0389-8 pmid: 30988512 |
| [33] | Betsuyaku S, Sawa S, Yamada M. The function of the CLE peptides in plant development and plant-microbe interactions[J]. Arabidopsis Book, 2011, 9: e0149. |
| [34] | Zhang LH, Yang Y, Mu CQ, et al. Control of root stem cell differentiation and lateral root emergence by CLE16/17 peptides in Arabidopsis[J]. Front Plant Sci, 2022, 13: 869888. |
| [35] | Gregory EF, Dao TQ, Alexander MA, et al. The signaling peptide-encoding genes CLE16, CLE17 and CLE27 are dispensable for Arabidopsis shoot apical meristem activity[J]. PLoS One, 2018, 13(8): e0202595. |
| [36] | Kondo T, Sawa S, Kinoshita A, et al. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis[J]. Science, 2006, 313(5788): 845-848. |
| [37] | Ohyama K, Shinohara H, Ogawa-Ohnishi M, et al. A glycopeptide regulating stem cell fate in Arabidopsis thaliana[J]. Nat Chem Biol, 2009, 5(8): 578-580. |
| [38] | Shinohara H, Matsubayashi Y. Reevaluation of the CLV3-receptor interaction in the shoot apical meristem: dissection of the CLV3 signaling pathway from a direct ligand-binding point of view[J]. Plant J, 2015, 82(2): 328-336. |
| [39] | Kim HJ, Wu CY, Yu HM, et al. Dual CLAVATA3 peptides in Arabidopsis shoot stem cell signaling[J]. J Plant Biol, 2017, 60(5): 506-512. |
| [40] | Shinohara H, Moriyama Y, Ohyama K, et al. Biochemical mapping of a ligand-binding domain within Arabidopsis BAM1 reveals diversified ligand recognition mechanisms of plant LRR-RKs[J]. Plant J, 2012, 70(5): 845-854. |
| [41] | Song XF, Yu DL, Xu TT, et al. Contributions of individual amino acid residues to the endogenous CLV3 function in shoot apical meristem maintenance in Arabidopsis[J]. Mol Plant, 2012, 5(2): 515-523. |
| [42] | MacAlister CA, Ortiz-Ramírez C, Becker JD, et al. Hydroxyproline O-arabinosyltransferase mutants oppositely alter tip growth in Arabidopsis thaliana and Physcomitrella patens[J]. Plant J, 2016, 85(2): 193-208. |
| [43] |
Xu C, Liberatore KL, MacAlister CA, et al. A cascade of Arabinosyltransferases controls shoot meristem size in tomato[J]. Nat Genet, 2015, 47(7): 784-792.
doi: 10.1038/ng.3309 pmid: 26005869 |
| [44] |
Dievart A, Gottin C, Périn C, et al. Origin and diversity of plant receptor-like kinases[J]. Annu Rev Plant Biol, 2020, 71: 131-156.
doi: 10.1146/annurev-arplant-073019-025927 pmid: 32186895 |
| [45] |
Clark SE, Williams RW, Meyerowitz EM. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis[J]. Cell, 1997, 89(4): 575-585.
doi: 10.1016/s0092-8674(00)80239-1 pmid: 9160749 |
| [46] |
Bleckmann A, Weidtkamp-Peters S, Seidel CAM, et al. Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane[J]. Plant Physiol, 2010, 152(1): 166-176.
doi: 10.1104/pp.109.149930 pmid: 19933383 |
| [47] | Zhu YF, Wang YQ, Li RL, et al. Analysis of interactions among the CLAVATA3 receptors reveals a direct interaction between CLAVATA2 and CORYNE in Arabidopsis[J]. Plant J, 2010, 61(2): 223-233. |
| [48] |
Jeong S, Trotochaud AE, Clark SE. The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase[J]. Plant Cell, 1999, 11(10): 1925-1934.
doi: 10.1105/tpc.11.10.1925 pmid: 10521522 |
| [49] |
Müller R, Bleckmann A, Simon R. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1[J]. Plant Cell, 2008, 20(4): 934-946.
doi: 10.1105/tpc.107.057547 pmid: 18381924 |
| [50] | Nimchuk ZL, Tarr PT, Meyerowitz EM. An evolutionarily conserved pseudokinase mediates stem cell production in plants[J]. Plant Cell, 2011, 23(3): 851-854. |
| [51] |
Kinoshita A, Betsuyaku S, Osakabe Y, et al. RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis[J]. Development, 2010, 137(22): 3911-3920.
doi: 10.1242/dev.048199 pmid: 20978082 |
| [52] |
Hu C, Zhu YF, Cui YW, et al. A group of receptor kinases are essential for CLAVATA signalling to maintain stem cell homeostasis[J]. Nat Plants, 2018, 4(4): 205-211.
doi: 10.1038/s41477-018-0123-z pmid: 29581511 |
| [53] | Wang WP, Hu C, Li XN, et al. Receptor-like cytoplasmic kinases PBL34/35/36 are required for CLE peptide-mediated signaling to maintain shoot apical meristem and root apical meristem homeostasis in Arabidopsis[J]. Plant Cell, 2022, 34(4): 1289-1307. |
| [54] |
Betsuyaku S, Takahashi F, Kinoshita A, et al. Mitogen-activated protein kinase regulated by the CLAVATA receptors contributes to shoot apical meristem homeostasis[J]. Plant Cell Physiol, 2011, 52(1): 14-29.
doi: 10.1093/pcp/pcq157 pmid: 20965998 |
| [55] | Chou H, Zhu YF, Ma Y, et al. The CLAVATA signaling pathway mediating stem cell fate in shoot meristems requires Ca(2+)as a secondary cytosolic messenger[J]. Plant J, 2016, 85(4): 494-506. |
| [56] | Williams RW, Wilson JM, Meyerowitz EM. A possible role for kinase-associated protein phosphatase in the Arabidopsis CLAVATA1 signaling pathway[J]. Proc Natl Acad Sci U S A, 1997, 94(19): 10467-10472. |
| [57] | Yu LP, Miller AK, Clark SE. POLTERGEIST encodes a protein phosphatase 2C that regulates CLAVATA pathways controlling stem cell identity at Arabidopsis shoot and flower meristems[J]. Curr Biol, 2003, 13(3): 179-188. |
| [58] | Song SK, Lee MM, Clark SE. POL and PLL1 phosphatases are CLAVATA1 signaling intermediates required for Arabidopsis shoot and floral stem cells[J]. Development, 2006, 133(23): 4691-4698. |
| [59] |
Trotochaud AE, Hao T, Wu G, et al. The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a Rho-related protein[J]. Plant Cell, 1999, 11(3): 393-406.
doi: 10.1105/tpc.11.3.393 pmid: 10072399 |
| [60] | Bommert P, Je BI, Goldshmidt A, et al. The maize Gα gene COMPACT PLANT2 functions in CLAVATA signalling to control shoot meristem size[J]. Nature, 2013, 502(7472): 555-558. |
| [61] | Je BI, Xu F, Wu QY, et al. The CLAVATA receptor FASCIATED EAR2 responds to distinct CLE peptides by signaling through two downstream effectors[J]. eLife, 2018, 7: e35673. |
| [62] | Ishida T, Tabata R, Yamada M, et al. Heterotrimeric G proteins control stem cell proliferation through CLAVATA signaling in Arabidopsis[J]. EMBO Rep, 2014, 15(11): 1202-1209. |
| [63] |
Shimada T, Sugano SS, Hara-Nishimura I. Positive and negative peptide signals control stomatal density[J]. Cell Mol Life Sci, 2011, 68(12): 2081-2088.
doi: 10.1007/s00018-011-0685-7 pmid: 21509541 |
| [64] | Shinohara H. Root meristem growth factor RGF, a sulfated peptide hormone in plants[J]. Peptides, 2021, 142: 170556. |
| [65] |
Furumizu C, Aalen RB. Peptide signaling through leucine-rich repeat receptor kinases: insight into land plant evolution[J]. New Phytol, 2023, 238(3): 977-982.
doi: 10.1111/nph.18827 pmid: 36811171 |
| [66] |
Rowe MH, Bergmann DC. Complex signals for simple cells: the expanding ranks of signals and receptors guiding stomatal development[J]. Curr Opin Plant Biol, 2010, 13(5): 548-555.
doi: 10.1016/j.pbi.2010.06.002 pmid: 20638894 |
| [67] |
Kondo T, Kajita R, Miyazaki A, et al. Stomatal density is controlled by a mesophyll-derived signaling molecule[J]. Plant Cell Physiol, 2010, 51(1): 1-8.
doi: 10.1093/pcp/pcp180 pmid: 20007289 |
| [68] |
Kosentka PZ, Overholt A, Maradiaga R, et al. EPFL signals in the boundary region of the SAM restrict its size and promote leaf initiation[J]. Plant Physiol, 2019, 179(1): 265-279.
doi: 10.1104/pp.18.00714 pmid: 30409857 |
| [69] |
Uchida N, Shimada M, Tasaka M. ERECTA-family receptor kinases regulate stem cell homeostasis via buffering its cytokinin responsiveness in the shoot apical meristem[J]. Plant Cell Physiol, 2013, 54(3): 343-351.
doi: 10.1093/pcp/pcs109 pmid: 22885615 |
| [70] | Zhang L, DeGennaro D, Lin GZ, et al. ERECTA family signaling constrains CLAVATA3 and WUSCHEL to the center of the shoot apical meristem[J]. Development, 2021, 148(5): dev189753. |
| [71] |
Rellán-Álvarez R, Lobet G, Dinneny JR. Environmental control of root system biology[J]. Annu Rev Plant Biol, 2016, 67: 619-642.
doi: 10.1146/annurev-arplant-043015-111848 pmid: 26905656 |
| [72] |
Zhu YF, Hu C, Gou XP. Receptor-like protein kinase-mediated signaling in controlling root meristem homeostasis[J]. aBIOTECH, 2020, 1(3): 157-168.
doi: 10.1007/s42994-020-00024-z pmid: 36303569 |
| [73] | Zhu YF, Hu C, Cui YW, et al. Conserved and differentiated functions of CIK receptor kinases in modulating stem cell signaling in Arabidopsis[J]. Mol Plant, 2021, 14(7): 1119-1134. |
| [74] | Stahl Y, Wink RH, Ingram GC, et al. A signaling module controlling the stem cell niche in Arabidopsis root meristems[J]. Curr Biol, 2009, 19(11): 909-914. |
| [75] | Stahl Y, Grabowski S, Bleckmann A, et al. Moderation of Arabidop- sis root stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 receptor kinase complexes[J]. Curr Biol, 2013, 23(5): 362-371. |
| [76] | Sarkar AK, Luijten M, Miyashima S, et al. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers[J]. Nature, 2007, 446(7137): 811-814. |
| [77] | Ren SC, Song XF, Chen WQ, et al. CLE25 peptide regulates phloem initiation in Arabidopsis through a CLERK-CLV2 receptor complex[J]. J Integr Plant Biol, 2019, 61(10): 1043-1061. |
| [78] |
Rodriguez-Villalon A. Wiring a plant: genetic networks for phloem formation in Arabidopsis thaliana roots[J]. New Phytol, 2016, 210(1): 45-50.
doi: 10.1111/nph.13527 pmid: 26171671 |
| [79] |
Hazak O, Brandt B, Cattaneo P, et al. Perception of root-active CLE peptides requires CORYNE function in the phloem vasculature[J]. EMBO Rep, 2017, 18(8): 1367-1381.
doi: 10.15252/embr.201643535 pmid: 28607033 |
| [80] | Anne P, Amiguet-Vercher A, Brandt B, et al. CLERK is a novel receptor kinase required for sensing of root-active CLE peptides in Arabidopsis[J]. Development, 2018, 145(10): dev162354. |
| [81] | Depuydt S, Rodriguez-Villalon A, Santuari L, et al. Suppression of Arabidopsis protophloem differentiation and root meristem growth by CLE45 requires the receptor-like kinase BAM3[J]. Proc Natl Acad Sci USA, 2013, 110(17): 7074-7079. |
| [82] | Hu C, Zhu YF, Cui YW, et al. A CLE-BAM-CIK signalling module controls root protophloem differentiation in Arabidopsis[J]. New Phytol, 2022, 233(1): 282-296. |
| [83] |
Shimizu N, Ishida T, Yamada M, et al. Bam 1 and receptor-like protein kinase 2 constitute a signaling pathway and modulate cle peptide-triggered growth inhibition in Arabidopsis root[J]. New Phytol, 2015, 208(4): 1104-1113.
doi: 10.1111/nph.13520 pmid: 26083273 |
| [84] |
Gujas B, Kastanaki E, Sturchler A, et al. A reservoir of pluripotent phloem cells safeguards the linear developmental trajectory of protophloem sieve elements[J]. Curr Biol, 2020, 30(5): 755-766.e4.
doi: S0960-9822(19)31676-8 pmid: 32037095 |
| [85] |
Breda AS, Hazak O, Schultz P, et al. A cellular insulator against CLE45 peptide signaling[J]. Curr Biol, 2019, 29(15): 2501-2508.e3.
doi: S0960-9822(19)30764-X pmid: 31327718 |
| [86] |
Rodriguez-Villalon A, Gujas B, Kang YH, et al. Molecular genetic framework for protophloem formation[J]. Proc Natl Acad Sci USA, 2014, 111(31): 11551-11556.
doi: 10.1073/pnas.1407337111 pmid: 25049386 |
| [87] |
Wang Q, Aliaga Fandino AC, Graeff M, et al. A phosphoinositide hub connects CLE peptide signaling and polar auxin efflux regulation[J]. Nat Commun, 2023, 14(1): 423.
doi: 10.1038/s41467-023-36200-0 pmid: 36702874 |
| [88] | Kang YH, Hardtke CS. Arabidopsis MAKR5 is a positive effector of BAM3-dependent CLE45 signaling[J]. EMBO Rep, 2016, 17(8): 1145-1154. |
| [89] | Diaz-Ardila HN, Gujas B, Wang Q, et al. pH-dependent CLE peptide perception permits phloem differentiation in Arabidopsis roots[J]. Curr Biol, 2023, 33(3): 597-605.e3. |
| [90] |
Whitford R, Fernandez A, Tejos R, et al. GOLVEN secretory peptides regulate auxin carrier turnover during plant gravitropic responses[J]. Dev Cell, 2012, 22(3): 678-685.
doi: 10.1016/j.devcel.2012.02.002 pmid: 22421050 |
| [91] |
Matsuzaki Y, Ogawa-Ohnishi M, Mori A, et al. Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis[J]. Science, 2010, 329(5995): 1065-1067.
doi: 10.1126/science.1191132 pmid: 20798316 |
| [92] | Meng L, Buchanan BB, Feldman LJ, et al. CLE-like(CLEL)peptides control the pattern of root growth and lateral root development in Arabidopsis[J]. Proc Natl Acad Sci USA, 2012, 109(5): 1760-1765. |
| [93] | Ou Y, Lu XT, Zi QE, et al. RGF1 INSENSITIVE 1 to 5, a group of LRR receptor-like kinases, are essential for the perception of root meristem growth factor 1 in Arabidopsis thaliana[J]. Cell Res, 2016, 26(6): 686-698. |
| [94] |
Song W, Liu L, Wang JZ, et al. Signature motif-guided identification of receptors for peptide hormones essential for root meristem growth[J]. Cell Res, 2016, 26(6): 674-685.
doi: 10.1038/cr.2016.62 pmid: 27229311 |
| [95] |
Komori R, Amano Y, Ogawa-Ohnishi M, et al. Identification of tyrosylprotein sulfotransferase in Arabidopsis[J]. Proc Natl Acad Sci USA, 2009, 106(35): 15067-15072.
doi: 10.1073/pnas.0902801106 pmid: 19666544 |
| [96] |
Shinohara H, Mori A, Yasue N, et al. Identification of three LRR-RKs involved in perception of root meristem growth factor in Arabidopsis[J]. Proc Natl Acad Sci USA, 2016, 113(14): 3897-3902.
doi: 10.1073/pnas.1522639113 pmid: 27001831 |
| [97] | Aida M, Beis D, Heidstra R, et al. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche[J]. Cell, 2004, 119(1): 109-120. |
| [98] | Galinha C, Hofhuis H, Luijten M, et al. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development[J]. Nature, 2007, 449(7165): 1053-1057. |
| [99] | Mähönen AP, Ten Tusscher K, Siligato R, et al. PLETHORA gradient formation mechanism separates auxin responses[J]. Nature, 2014, 515(7525): 125-129. |
| [100] | Shimotohno A, Heidstra R, Blilou I, et al. Root stem cell niche organizer specification by molecular convergence of PLETHORA and SCARECROW transcription factor modules[J]. Genes Dev, 2018, 32(15-16): 1085-1100. |
| [101] | Yamada M, Han XW, Benfey PN. RGF1 controls root meristem size through ROS signalling[J]. Nature, 2020, 577(7788): 85-88. |
| [102] |
Shao YM, Yu XX, Xu XW, et al. The YDA-MKK4/MKK5-MPK3/MPK6 cascade functions downstream of the RGF1-RGI ligand-receptor pair in regulating mitotic activity in root apical meristem[J]. Mol Plant, 2020, 13(11): 1608-1623.
doi: 10.1016/j.molp.2020.09.004 pmid: 32916336 |
| [103] | Lu XT, Shi HY, Ou Y, et al. RGF1-RGI1, a peptide-receptor complex, regulates Arabidopsis root meristem development via a MAPK signaling cascade[J]. Mol Plant, 2020, 13(11): 1594-1607. |
| [104] | Zhou WK, Wei LR, Xu J, et al. Arabidopsis Tyrosylprotein sulfotransferase acts in the auxin/PLETHORA pathway in regulating postembryonic maintenance of the root stem cell niche[J]. Plant Cell, 2010, 22(11): 3692-3709. |
| [105] | Takahashi F, Suzuki T, Osakabe Y, et al. A small peptide modulates stomatal control via abscisic acid in long-distance signalling[J]. Nature, 2018, 556(7700): 235-238. |
| [106] |
Araya T, Miyamoto M, Wibowo J, et al. CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner[J]. Proc Natl Acad Sci USA, 2014, 111(5): 2029-2034.
doi: 10.1073/pnas.1319953111 pmid: 24449877 |
| [107] |
Sánchez-Calderón L, López-Bucio J, Chacón-López A, et al. Phosphate starvation induces a determinate developmental program in the roots of Arabidopsis thaliana[J]. Plant Cell Physiol, 2005, 46(1): 174-184.
doi: 10.1093/pcp/pci011 pmid: 15659445 |
| [108] |
Gutiérrez-Alanís D, Yong-Villalobos L, Jiménez-Sandoval P, et al. Phosphate starvation-dependent iron mobilization induces CLE14 expression to trigger root meristem differentiation through CLV2/PEPR2 signaling[J]. Dev Cell, 2017, 41(5): 555-570.e3.
doi: S1534-5807(17)30391-X pmid: 28586647 |
| [109] |
Cederholm HM, Benfey PN. Distinct sensitivities to phosphate deprivation suggest that RGF peptides play disparate roles in Arabidopsis thaliana root development[J]. New Phytol, 2015, 207(3): 683-691.
doi: 10.1111/nph.13405 pmid: 25856240 |
| [110] |
Long J, Barton MK. Initiation of axillary and floral meristems in Arabidopsis[J]. Dev Biol, 2000, 218(2): 341-353.
doi: 10.1006/dbio.1999.9572 pmid: 10656774 |
| [111] |
Wu QY, Xu F, Jackson D. All together now, a magical mystery tour of the maize shoot meristem[J]. Curr Opin Plant Biol, 2018, 45(Pt A): 26-35.
doi: S1369-5266(18)30024-4 pmid: 29778985 |
| [112] | Liu L, Gallagher J, Arevalo ED, et al. Enhancing grain-yield-related traits by CRISPR-Cas9 promoter editing of maize CLE genes[J]. Nat Plants, 2021, 7(3): 287-294. |
| [113] | Chu HW, Qian Q, Liang WQ, et al. The floral organ number4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice[J]. Plant Physiol, 2006, 142(3): 1039-1052. |
| [114] | Suzaki T, Toriba T, Fujimoto M, et al. Conservation and diversification of meristem maintenance mechanism in Oryza sativa: function of the FLORAL ORGAN NUMBER2 gene[J]. Plant Cell Physiol, 2006, 47(12): 1591-1602. |
| [115] | Ohmori Y, Tanaka W, Kojima M, et al. WUSCHEL-RELATED HOMEOBOX4 is involved in meristem maintenance and is negatively regulated by the CLE gene FCP1 in rice[J]. Plant Cell, 2013, 25(1): 229-241. |
| [116] | 吴琳华, 陈文丰. 水稻花序结构调控机制研究进展[J]. 广东农业科学, 2022, 49(9): 42-52. |
| Wu LH, Chen WF. Research progress in regulation mechanism of rice inflorescence structure[J]. Guangdong Agric Sci, 2022, 49(9): 42-52. | |
| [117] | Suzaki T, Sato M, Ashikari M, et al. The gene FLORAL ORGAN NUMBER1 regulates floral meristem size in rice and encodes a leucine-rich repeat receptor kinase orthologous to Arabidopsis CLAVATA1[J]. Development, 2004, 131(22): 5649-5657. |
| [118] | Suzaki T, Ohneda M, Toriba T, et al. FON2 SPARE1 redundantly regulates floral meristem maintenance with FLORAL ORGAN NUMBER2 in rice[J]. PLoS Genet, 2009, 5(10): e1000693. |
| [119] |
Suzaki T, Yoshida A, Hirano HY. Functional diversification of CLAVATA3-related CLE proteins in meristem maintenance in rice[J]. Plant Cell, 2008, 20(8): 2049-2058.
doi: 10.1105/tpc.107.057257 pmid: 18676878 |
| [120] |
Satterlee JW, Strable J, Scanlon MJ. Plant stem-cell organization and differentiation at single-cell resolution[J]. Proc Natl Acad Sci USA, 2020, 117(52): 33689-33699.
doi: 10.1073/pnas.2018788117 pmid: 33318187 |
| [121] |
Knauer S, Javelle M, Li L, et al. A high-resolution gene expression atlas links dedicated meristem genes to key architectural traits[J]. Genome Res, 2019, 29(12): 1962-1973.
doi: 10.1101/gr.250878.119 pmid: 31744902 |
| [122] |
Je BI, Gruel J, Lee YK, et al. Signaling from maize organ primordia via FASCIATED EAR3 regulates stem cell proliferation and yield traits[J]. Nat Genet, 2016, 48(7): 785-791.
doi: 10.1038/ng.3567 pmid: 27182966 |
| [123] |
Bommert P, Nagasawa NS, Jackson D. Quantitative variation in maize kernel row number is controlled by the FASCIATED EAR2 locus[J]. Nat Genet, 2013, 45(3): 334-337.
doi: 10.1038/ng.2534 pmid: 23377180 |
| [124] | Taguchi-Shiobara F, Yuan Z, Hake S, et al. The fasciated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize[J]. Genes Dev, 2001, 15(20): 2755-2766. |
| [125] |
Bommert P, Lunde CN, Nardmann J, et al. Thick tassel dwarf1 encodes a putative maize ortholog of the Arabidopsis CLAVATA1 leucine-rich repeat receptor-like kinase[J]. Development, 2005, 132(6): 1235-1245.
doi: 10.1242/dev.01671 pmid: 15716347 |
| [126] | Kwon CT, Tang LL, Wang XG, et al. Dynamic evolution of small signalling peptide compensation in plant stem cell control[J]. Nat Plants, 2022, 8(4): 346-355. |
| [127] |
Wolter F, Schindele P, Puchta H. Plant breeding at the speed of light: the power of CRISPR/Cas to generate directed genetic diversity at multiple sites[J]. BMC Plant Biol, 2019, 19(1): 176.
doi: 10.1186/s12870-019-1775-1 pmid: 31046670 |
| [128] |
Rodríguez-Leal D, Lemmon ZH, Man J, et al. Engineering quantitative trait variation for crop improvement by genome editing[J]. Cell, 2017, 171(2): 470-480.e8.
doi: S0092-8674(17)30988-1 pmid: 28919077 |
| [129] | Jin J, Hua L, Zhu ZF, et al. GAD1 encodes a secreted peptide that regulates grain number, grain length, and awn development in rice domestication[J]. Plant Cell, 2016, 28(10): 2453-2463. |
| [130] | Jin J, Xiong LL, Gray JE, et al. Two awn-development-related peptides, GAD1 and OsEPFL2, promote seed dispersal and germination in rice[J]. Mol Plant, 2023, 16(3): 485-488. |
| [131] |
Guo T, Lu ZQ, Xiong YH, et al. Optimization of rice panicle architecture by specifically suppressing ligand-receptor pairs[J]. Nat Commun, 2023, 14(1): 1640.
doi: 10.1038/s41467-023-37326-x pmid: 36964129 |
| [132] |
Muños S, Ranc N, Botton E, et al. Increase in tomato locule number is controlled by two single-nucleotide polymorphisms located near WUSCHEL[J]. Plant Physiol, 2011, 156(4): 2244-2254.
doi: 10.1104/pp.111.173997 pmid: 21673133 |
| [133] | Lee H. Stem cell maintenance and abiotic stress response in shoot apical meristem for developmental plasticity[J]. J Plant Biol, 2018, 61(6): 358-365. |
| [134] | Wang SX, Tian L, Liu HJ, et al. Large-scale discovery of non-conventional peptides in maize and Arabidopsis through an integrated peptidogenomic pipeline[J]. Mol Plant, 2020, 13(7): 1078-1093. |
| [135] | Khaipho-Burch M, Cooper M, Crossa J, et al. Genetic modification can improve crop yields—but stop overselling it[J]. Nature, 2023, 621: 470-473. |
| [1] | WANG Di ZHANG Xiao-yu SONG Yu-xin ZHENG Dong-ran TIAN Jing LI Yu-hua WANG Yu WU Hao. Advances in the Molecular Mechanisms of Plant Tissue Culture and Regeneration Regulated by Totipotency-related Transcription Factors [J]. Biotechnology Bulletin, 2024, 40(6): 23-33. |
| [2] | LU Zhen-wan, LI Xue-qi, HUANG Jin-guang, ZHOU Huan-bin. Creation of Glyphosate-tolerant Rice by Cytosine Base Editing [J]. Biotechnology Bulletin, 2023, 39(2): 63-69. |
| [3] | LI Peng-cheng, ZHANG Ming-jun, WANG Yin-xiao, LI Xiang-yin, LI Sheng-yan, LANG Zhi-hong. Insect Resistance Identification and Agronomy Traits Analysis of Transgenic Maize HGK60 with Different Genetic Backgrounds [J]. Biotechnology Bulletin, 2023, 39(1): 40-47. |
| [4] | WANG Cui-cui, FU Da-qi. Research Progress in the Effects of Ubiquitin-proteasome System on Plant Agronomic Traits [J]. Biotechnology Bulletin, 2023, 39(1): 72-83. |
| [5] | ZHANG Chan, WU You-gen, YU Jing, YANG Dong-mei, YAO Guang-long, YANG Hua-geng, ZHANG Jun-feng, CHEN Ping. Molecular Mechanism of Terpenoids Synthesis Intermediated by Light and Jasmonates Signals [J]. Biotechnology Bulletin, 2022, 38(8): 32-40. |
| [6] | ZHANG Feng, CHEN Wei. Research Progress of Metabolomics in Plant Stress Biology [J]. Biotechnology Bulletin, 2021, 37(8): 1-11. |
| [7] | QIAN Hong-ping, CHEN Bo, LIN Jin-xing, CUI Ya-ning. Recent Advances on Dynamic Regulation and Imaging Techniques of RNA Polymerase II [J]. Biotechnology Bulletin, 2021, 37(4): 293-302. |
| [8] | LI Ze-qing, LIU Cai-xian, XING Wen, WEN Ya-feng. Research Progress on Regulation of miRNA in the Heat Stress Response of Plants [J]. Biotechnology Bulletin, 2020, 36(2): 149-157. |
| [9] | ZHENG Wen-qing, ZHANG Qian, DU Liang. Short Tandem Target Mimic and Its Application in Analyzing Plant miRNA Functions [J]. Biotechnology Bulletin, 2020, 36(12): 256-264. |
| [10] | CHANG Yong-fang, BAO Peng-jia, CHU Min, WU Xiao-yun, LIANG Chun-nian, YAN Ping. Research Progress on the Regulation of LncRNA in the Development of Mammalian Hair Follicle [J]. Biotechnology Bulletin, 2019, 35(8): 205-212. |
| [11] | MO Xian-lan, SHI Lie-qin, LU Qiu-li, WANG Xiao-min, REN Zhen-xin. Expression Analysis of Sl-miR482 in Tomato Fruit and the Construction of STTM Silencing Vector [J]. Biotechnology Bulletin, 2019, 35(12): 50-56. |
| [12] | HUANG Xing, DING Feng, PENG Hong-xiang, PAN Jie-chun, HE Xin-hua, XU Jiong-zhi, LI Lin. Research Progress on Family of Plant WRKY Transcription Factors [J]. Biotechnology Bulletin, 2019, 35(12): 129-143. |
| [13] | LIANG Hai-sheng, LI Meng-tao, LI Sheng-yan, WANG Hai, ZHANG Jie, LANG Zhi-hong. Agronomic Traits Analysis of Transgenic Bt cry1Ah Maize HGK60 Line [J]. Biotechnology Bulletin, 2018, 34(7): 92-100. |
| [14] | TAN Yu-rong, WANG Dan, GAO Xuan, LIU Jin-ping. Research Advance on Plant Long Noncoding RNAs [J]. Biotechnology Bulletin, 2018, 34(10): 1-10. |
| [15] | SONG Yan-chao, An Fei-fei, Xue Jing-jing, Qin Yu-ling, Li Kai-mian, CHEN Song-bi. Proteomic Analysis on Tuberous Roots of Cassava Cultivar ZM-Seaside and Mosaic-leaf Mutation [J]. Biotechnology Bulletin, 2017, 33(3): 78-85. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||