








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (7): 125-136.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0132
Previous Articles Next Articles
PANG Meng-zhen1,2( ), XU Han-qin1,2, LIU Hai-yan1,2, SONG Juan1,2, WANG Jia-han1,2, SUN Li-na1,2, JI Pei-mei1,2, YIN Ze-zhi1,2, HU You-chuan1,2, ZHAO Xiao-meng1,2, LIANG Shan-shan1,2, ZHANG Si-ju1,2(
), XU Han-qin1,2, LIU Hai-yan1,2, SONG Juan1,2, WANG Jia-han1,2, SUN Li-na1,2, JI Pei-mei1,2, YIN Ze-zhi1,2, HU You-chuan1,2, ZHAO Xiao-meng1,2, LIANG Shan-shan1,2, ZHANG Si-ju1,2( ), LUAN Wei-jiang1,2(
), LUAN Wei-jiang1,2( )
)
Received:2024-02-02
Online:2024-07-26
Published:2024-05-24
Contact:
ZHANG Si-ju, LUAN Wei-jiang
E-mail:mengzhenpang@163.com;zhangsiju@126.com;lwjzsq@163.com
PANG Meng-zhen, XU Han-qin, LIU Hai-yan, SONG Juan, WANG Jia-han, SUN Li-na, JI Pei-mei, YIN Ze-zhi, HU You-chuan, ZHAO Xiao-meng, LIANG Shan-shan, ZHANG Si-ju, LUAN Wei-jiang. Gene Identification and Functional Analysis of Yellowish and Early Heading Mutant hz1 in Rice[J]. Biotechnology Bulletin, 2024, 40(7): 125-136.
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 内切酶 Endonuclease |
|---|---|---|
| SE5-GFP-F | GTTGGTACCCGCTATAAGAGGGAGAGAGG | Kpn Ⅰ |
| SE5-GFP-R | CCGTCTAGAGGTGAATATGTGACGGAGGA | Xba Ⅰ |
| LB-R | ACGATGGACTCCAGTCCGGCCcttgaccaactctatcagagcttgg | |
| HZ1-F | CAAGGACCAGGCCAAGGAAG | |
| HZ1-R | ACCAAACTCAAAACAGGGGG | |
| InDel1-F | CAGGTATTCTGAGGTTGTATCC | |
| InDel1-R | CACGGTCAAATTCAACATTCC | |
| dCAP3-F | CACCAACTGAGCTCACTAGCCATAT | NdeⅠ |
| dCAP3-R | GGCCCAATGACTACCTTCTACTTTA | |
| SE5-ex-F | GGCTGAAAAGGACTCCCAAG | |
| SE5-ex-R | GTCCAGCTAGAGGCGACTTC | |
| Actin1-ex-F | GACTCTGGTGATGGTGTCAGC | |
| Actin1-ex-R | GGCTGGAAGAGGACCTCAGG | |
| Hd3a-ex-F | TTGGTAGGGTTGTGGGTGATGTGC | |
| Hd3a-ex-R | AGGTTAGGGTCACTTGGGCTTGGT | |
| RFT1-ex-F | TCCGAGCCCAAGCAACCCTAAC | |
| RFT1-ex-R | AGTTCCTGGTGCTGAAGTTCTG | |
| HEMA-ex-F | GATGCAATCACTGCTGGAAAGCGT | |
| HEMA-ex-R | CCATCTTGCCAGCACCAATCAACA | |
| PORA-ex-F | TCGTCGGCCTCGTCTGAGTTTATT | |
| PORA-ex-R | AGGCCTCTCTCACTGAAAGCTGAA | |
| YGL1-ex-F | CCAGCCACTGATGAAAGCAGCAAT | |
| YGL1-ex-R | AGAGCGCTAATACACTCGCGAACA | |
| CAO1-ex-F | CTTGTCGTATTCTTGGCGAG | |
| CAO1-ex-R | ATCCCGTGATGCTGTCGCTA |
Table 1 Primer sequence
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 内切酶 Endonuclease |
|---|---|---|
| SE5-GFP-F | GTTGGTACCCGCTATAAGAGGGAGAGAGG | Kpn Ⅰ |
| SE5-GFP-R | CCGTCTAGAGGTGAATATGTGACGGAGGA | Xba Ⅰ |
| LB-R | ACGATGGACTCCAGTCCGGCCcttgaccaactctatcagagcttgg | |
| HZ1-F | CAAGGACCAGGCCAAGGAAG | |
| HZ1-R | ACCAAACTCAAAACAGGGGG | |
| InDel1-F | CAGGTATTCTGAGGTTGTATCC | |
| InDel1-R | CACGGTCAAATTCAACATTCC | |
| dCAP3-F | CACCAACTGAGCTCACTAGCCATAT | NdeⅠ |
| dCAP3-R | GGCCCAATGACTACCTTCTACTTTA | |
| SE5-ex-F | GGCTGAAAAGGACTCCCAAG | |
| SE5-ex-R | GTCCAGCTAGAGGCGACTTC | |
| Actin1-ex-F | GACTCTGGTGATGGTGTCAGC | |
| Actin1-ex-R | GGCTGGAAGAGGACCTCAGG | |
| Hd3a-ex-F | TTGGTAGGGTTGTGGGTGATGTGC | |
| Hd3a-ex-R | AGGTTAGGGTCACTTGGGCTTGGT | |
| RFT1-ex-F | TCCGAGCCCAAGCAACCCTAAC | |
| RFT1-ex-R | AGTTCCTGGTGCTGAAGTTCTG | |
| HEMA-ex-F | GATGCAATCACTGCTGGAAAGCGT | |
| HEMA-ex-R | CCATCTTGCCAGCACCAATCAACA | |
| PORA-ex-F | TCGTCGGCCTCGTCTGAGTTTATT | |
| PORA-ex-R | AGGCCTCTCTCACTGAAAGCTGAA | |
| YGL1-ex-F | CCAGCCACTGATGAAAGCAGCAAT | |
| YGL1-ex-R | AGAGCGCTAATACACTCGCGAACA | |
| CAO1-ex-F | CTTGTCGTATTCTTGGCGAG | |
| CAO1-ex-R | ATCCCGTGATGCTGTCGCTA |
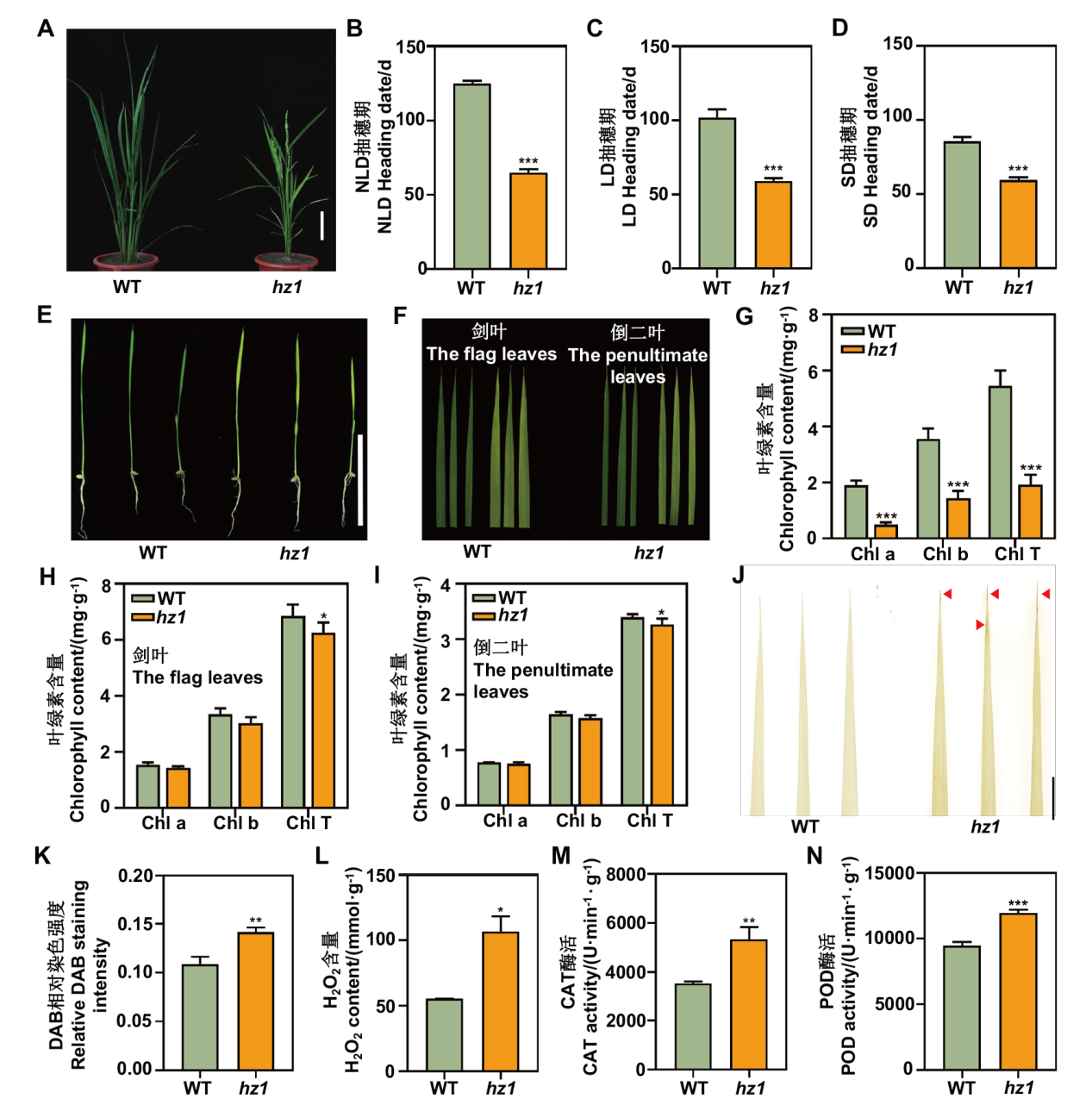
Fig. 1 Phenotype analysis of hz1 mutant A: The phenotype of hz1 mutant in the field, bar=10 cm. B-D: The heading dates of hz1 under natural long day(NLD), long day(LD), and short day(SD)conditions. Values are shown as mean ± SD(n ≥ 25). E: The phenotype of hz1 during the seedling stage, bar=1 cm. F: The leaves of hz1 at heading stage. G: Measurement of chlorophyll content during seedling stage. Values are shown as mean ± SD(n=3). H: The chlorophyll content of flag leaves during the heading stage. Values are shown as mean ± SD(n=3). I: The chlorophyll content of the penultimate leaves. Values are shown as mean ± SD(n=3)(Chl a: chlorophyll a; Chl b: chlorophyll b; Chl T: total chlorophyll content). J: The DAB staining results of WT and hz1. Red arrows indicate brown spots, bar=1 cm. K: Relative intensity of DAB staining for WT and hz1. Values are shown as mean ± SD(n=3). L: The measurement of hydrogen peroxide(H2O2)content in leaves. Values are shown as mean ± SD(n=3). M and N: Enzyme activity measurements of catalase(CAT)and peroxidase(POD)in leaves. Values are shown as mean ± SD(n=3). *: P<0.05; **: P<0.01; ***: P<0.001, the same below

Fig. 2 Analysis of agronomic traits of hz1 mutant A-G: Statistical analysis of plant height, tiller number, panicle length, number of primary branches, number of secondary branches, seed setting rate, and 100-grain weight. Values are shown as mean ± SD(n=30). H: The panicle of hz1, bar=10 cm. I: Grain number and seed setting rate of hz1, bar=10 cm. J and K: The pollen staining of hz1. The magnified times are 400 times, and the arrows indicate abortive pollens
| 组合 Cross | 正常表型植株数 Number of wild plants | 突变表型植株数 Number of mutant plants | 总植株数 Number of total plants | ꭕ2(3∶1) | P value |
|---|---|---|---|---|---|
| ZH11/hz1 | 458 | 130 | 588 | 2.621 3 | 0.105 4 |
Table 2 Genetic analysis of hz1 Mutant
| 组合 Cross | 正常表型植株数 Number of wild plants | 突变表型植株数 Number of mutant plants | 总植株数 Number of total plants | ꭕ2(3∶1) | P value |
|---|---|---|---|---|---|
| ZH11/hz1 | 458 | 130 | 588 | 2.621 3 | 0.105 4 |
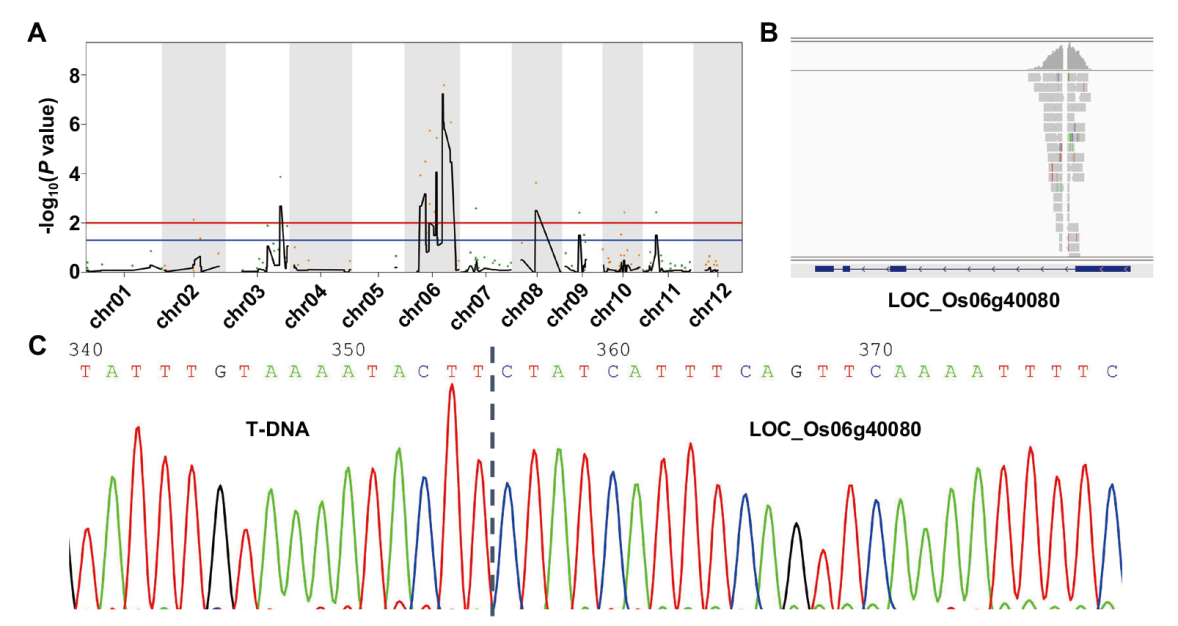
Fig. 3 BSA-seq analysis and gene mapping A: BSA-seq analysis. HZ1 is closely linked with the molecular marker on rice chromosome 6. B: T-DNA insertion site is co-segregated with HZ1. C: Sequencing analysis of T-DNA insertion site
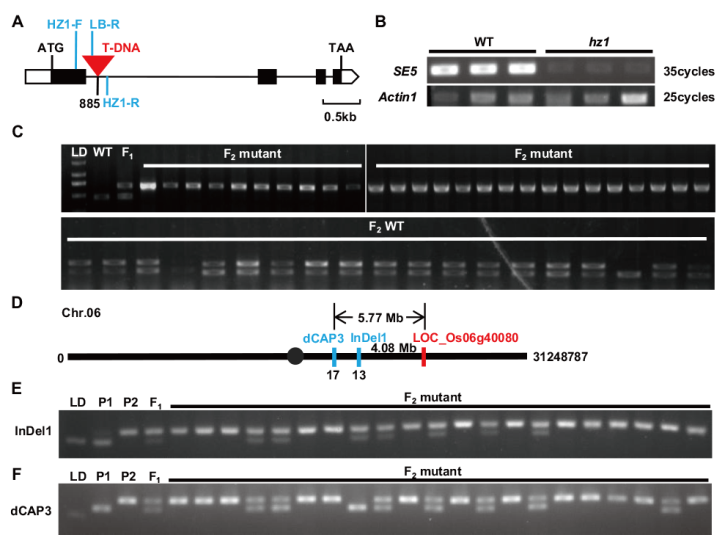
Fig. 4 Detection and linkage analysis of T-DNA insertion sites A: Schematic diagram of LOC_Os06g40080 site and T-DNA insertion location. HZ1-F, HZ1-R, and LB-R indicate the positions of the detection primers. B: RT-PCR analysis of HZ1/SE5 gene. Actin1 is an internal control in rice. C: The linkage analysis of T-DNA insertion sites. LD is DL 2000 marker, WT is wild-type, F1 is F1 plant, F2 mutant and F2 WT are plants with mutant phenotype and WT phenotype in the F2 population, respectively. D: Schematic diagram of InDel1, dCAP3 markers in the chromosome 6. E and F: PCR amplification of InDel1 and dCAP3. LD is DL 2000 marker, P1 is ZH11 plant, P2 is hz1 mutant, F1 is the F1 plants, and F2 mutant are the plants with hz1 phenotype in the F2 population
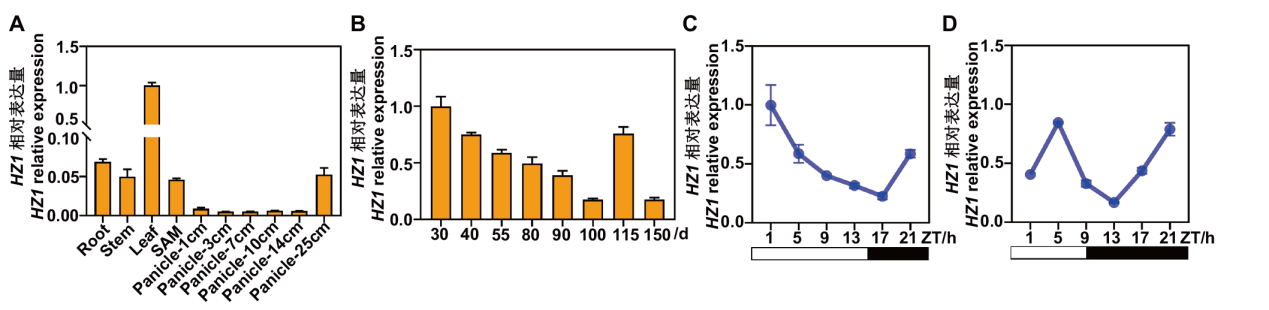
Fig. 5 Expression patterns of HZ1/SE5 gene A: Tissue-specific expression analysis of HZ1/SE5(SAM: stem apical meristem). B: Expression analysis of HZ1/SE5 at different growth stages in rice. C and D: Expression analysis of HZ1/SE5 under LD (C) and SD (D) conditions, respectively. ZT(zeitgeber time, h)indicates the time of light on. The white hollow box indicates the light period, and the black solid box indicates the dark period. The same below
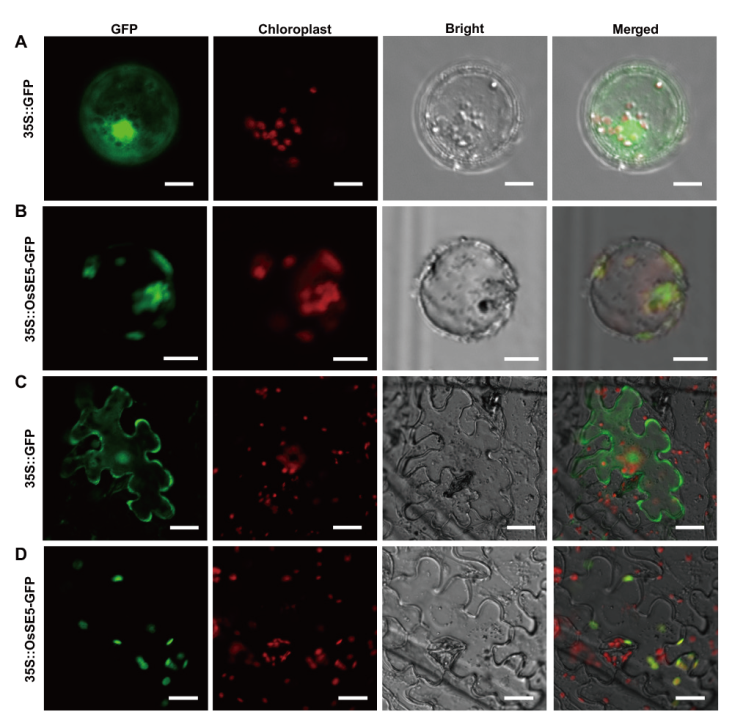
Fig. 6 Subcellular localization of HZ1/SE5 protein A and B: Transient expression of empty vector and HZ1-GFP fusion protein in rice protoplasts, bar=10 μm. C and D: Transient expression of empty vector and HZ1-GFP fusion protein in tobacco epidermal cells, bar=20 μm
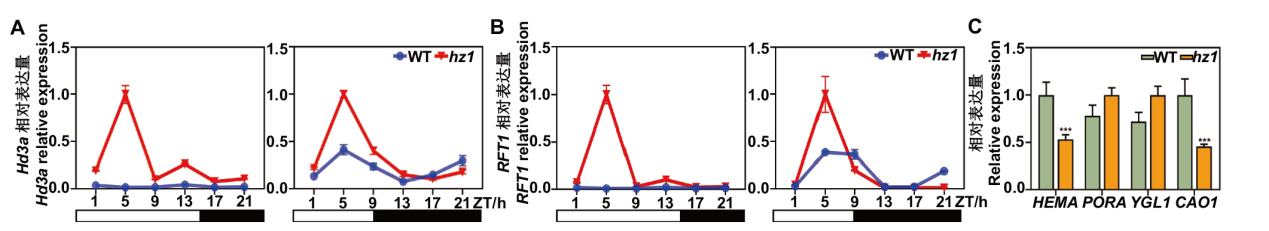
Fig. 7 Expression analysis of key genes involved in photoperiodic regulatory pathway and chlorophyll synthesis related genes in hz1 mutant A, B: Expression of key genes associated with photoperiodic regulatory pathway in WT and hz1 mutants under LD and SD conditions. C: Expression of chlorophyll synthesis related genes in WT and hz1 mutants
| [1] |
Yano M, Katayose Y, Ashikari M, et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS[J]. Plant Cell, 2000, 12(12): 2473-2484.
doi: 10.1105/tpc.12.12.2473 pmid: 11148291 |
| [2] | Lin HX, Yamamoto T, Sasaki T, et al. Characterization and detection of epistatic interactions of 3 QTLs, Hd1, Hd2, and Hd3, controlling heading date in rice using nearly isogenic lines[J]. Theor Appl Genet, 2000, 101(7): 1021-1028. |
| [3] |
Kojima S, Takahashi Y, Kobayashi Y, et al. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions[J]. Plant Cell Physiol, 2002, 43(10): 1096-1105.
doi: 10.1093/pcp/pcf156 pmid: 12407188 |
| [4] | Zhu YJ, Fan YY, Wang K, et al. Rice Flowering Locus T 1 plays an important role in heading date influencing yield traits in rice[J]. Sci Rep, 2017, 7(1): 4918. |
| [5] |
Komiya R, Ikegami A, Tamaki S, et al. Hd3a and RFT1 are essential for flowering in rice[J]. Development, 2008, 135(4): 767-774.
doi: 10.1242/dev.008631 pmid: 18223202 |
| [6] |
Xue WY, Xing YZ, Weng XY, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice[J]. Nat Genet, 2008, 40(6): 761-767.
doi: 10.1038/ng.143 pmid: 18454147 |
| [7] | Casal JJ, Luccioni LG, Oliverio KA, et al. Light, phytochrome signalling and photomorphogenesis in Arabidopsis[J]. Photochem Photobiol Sci, 2003, 2(6): 625-636. |
| [8] |
Wang X, Jiang BC, Gu LF, et al. A photoregulatory mechanism of the circadian clock in Arabidopsis[J]. Nat Plants, 2021, 7(10): 1397-1408.
doi: 10.1038/s41477-021-01002-z pmid: 34650267 |
| [9] |
Kami C, Lorrain S, Hornitschek P, et al. Light-regulated plant growth and development[J]. Curr Top Dev Biol, 2010, 91: 29-66.
doi: 10.1016/S0070-2153(10)91002-8 pmid: 20705178 |
| [10] |
Kliebenstein DJ, Lim JE, Landry LG, et al. Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1[J]. Plant Physiol, 2002, 130(1): 234-243.
doi: 10.1104/pp.005041 pmid: 12226503 |
| [11] | Cheng MC, Kathare PK, Paik I, et al. Phytochrome signaling networks[J]. Annu Rev Plant Biol, 2021, 72: 217-244. |
| [12] |
Shekhawat GS, Verma K. Haem oxygenase(HO): an overlooked enzyme of plant metabolism and defence[J]. J Exp Bot, 2010, 61(9): 2255-2270.
doi: 10.1093/jxb/erq074 pmid: 20378668 |
| [13] |
Davis SJ, Kurepa J, Vierstra RD. The Arabidopsis thaliana HY1 locus, required for phytochrome-chromophore biosynthesis, encodes a protein related to heme oxygenases[J]. Proc Natl Acad Sci USA, 1999, 96(11): 6541-6546.
doi: 10.1073/pnas.96.11.6541 pmid: 10339624 |
| [14] |
Izawa T, Oikawa T, Tokutomi S, et al. Phytochromes confer the photoperiodic control of flowering in rice(a short-day plant)[J]. Plant J, 2000, 22(5): 391-399.
doi: 10.1046/j.1365-313x.2000.00753.x pmid: 10849355 |
| [15] |
Andrés F, Galbraith DW, Talón M, et al. Analysis of PHOTOPERIOD SENSITIVITY5 sheds light on the role of phytochromes in photoperiodic flowering in rice[J]. Plant Physiol, 2009, 151(2): 681-690.
doi: 10.1104/pp.109.139097 pmid: 19675157 |
| [16] |
Chen H, Cheng ZJ, Ma XD, et al. A knockdown mutation of YELLOW-GREEN LEAF2 blocks chlorophyll biosynthesis in rice[J]. Plant Cell Rep, 2013, 32(12): 1855-1867.
doi: 10.1007/s00299-013-1498-y pmid: 24043333 |
| [17] | Peng YL, Zou T, Li LM, et al. Map-based cloning and functional analysis of YE1 in rice, which is involved in light-dependent chlorophyll biogenesis and photoperiodic flowering pathway[J]. Int J Mol Sci, 2019, 20(3): 758. |
| [18] | Rao YC, Xu N, Li SF, et al. PE-1, encoding heme oxygenase 1, impacts heading date and chloroplast development in rice(Oryza sativa L.)[J]. J Agric Food Chem, 2019, 67(26): 7249-7257. |
| [19] |
Arnon DI. Copper enzymes in isolated chloroplasts. polyphenoloxidase in beta vulgaris[J]. Plant Physiol, 1949, 24(1): 1-15.
doi: 10.1104/pp.24.1.1 pmid: 16654194 |
| [20] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T))Method[J]. Methods, 2001, 25(4): 402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [21] |
Trinidad JL, Longkumer T, Kohli A. Rice protoplast isolation and transfection for transient gene expression analysis[J]. Methods Mol Biol, 2021, 2238: 313-324.
doi: 10.1007/978-1-0716-1068-8_21 pmid: 33471341 |
| [22] | Parks BM, Quail PH. Phytochrome-deficient hy1 and hy2 long hypocotyl mutants of Arabidopsis are defective in phytochrome chromophore biosynthesis[J]. Plant Cell, 1991, 3(11): 1177-1186. |
| [23] |
Terry MJ, Kendrick RE. Feedback inhibition of chlorophyll synthesis in the phytochrome chromophore-deficient aurea and yellow-green-2 mutants of tomato[J]. Plant Physiol, 1999, 119(1): 143-152.
pmid: 9880355 |
| [24] | Kraepiel Y, Jullien M, Cordonnier-Pratt MM, et al. Identification of two loci involved in phytochrome expression in Nicotiana plumbaginifolia and lethality of the corresponding double mutant[J]. Mol Gen Genet, 1994, 242(5): 559-565. |
| [25] | Weller JL, Terry MJ, Rameau C, et al. The phytochrome-deficient pcd1 mutant of pea is unable to convert heme to biliverdin IXα[J]. Plant Cell, 1996, 8(1): 55-67. |
| [26] |
Davis SJ, Bhoo SH, Durski AM, et al. The heme-oxygenase family required for phytochrome chromophore biosynthesis is necessary for proper photomorphogenesis in higher plants[J]. Plant Physiol, 2001, 126(2): 656-669.
doi: 10.1104/pp.126.2.656 pmid: 11402195 |
| [27] | Legris M, Ince YÇ, Fankhauser C. Molecular mechanisms underlying phytochrome-controlled morphogenesis in plants[J]. Nat Commun, 2019, 10(1): 5219. |
| [28] |
Pham VN, Kathare PK, Huq E. Phytochromes and phytochrome interacting factors[J]. Plant Physiol, 2018, 176(2): 1025-1038.
doi: 10.1104/pp.17.01384 pmid: 29138351 |
| [29] |
Osugi A, Itoh H, Ikeda-Kawakatsu K, et al. Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice[J]. Plant Physiol, 2011, 157(3): 1128-1137.
doi: 10.1104/pp.111.181792 pmid: 21880933 |
| [30] | Wu HM, Zheng Y, Liu J, et al. Heme oxygenase-1 delays gibberellin-induced programmed cell death of rice aleurone layers subjected to drought stress by interacting with nitric oxide[J]. Front Plant Sci, 2016, 6: 1267. |
| [31] |
Hsu YY, Chao YY, Kao CH. Biliverdin-promoted lateral root formation is mediated through heme oxygenase in rice[J]. Plant Signal Behav, 2012, 7(7): 885-887.
doi: 10.4161/psb.20458 pmid: 22751314 |
| [32] | Xu S, Wang LJ, Zhang B, et al. RNAi knockdown of rice SE5 gene is sensitive to the herbicide methyl viologen by the down-regulation of antioxidant defense[J]. Plant Mol Biol, 2012, 80(2): 219-235. |
| [33] | Xie YJ, Mao Y, Xu S, et al. Heme-heme oxygenase 1 system is involved in ammonium tolerance by regulating antioxidant defence in Oryza sativa[J]. Plant Cell Environ, 2015, 38(1): 129-143. |
| [34] | Meng FY, Feng NJ, Zheng DF, et al. Exogenous Hemin alleviates NaCl stress by promoting photosynthesis and carbon metabolism in rice seedlings[J]. Sci Rep, 2023, 13(1): 3497. |
| [1] | SHEN Zhen-hui, CAO Yao, YANG Lin-lei, LUO Xiang-ying, ZI Ling-shan, LU Qing-qing, LI Rong-chun. Cloning and Bioinformatics Analysis of the Ergothioneine Biosynthesis Genes in Naematelia aurantialba and Stereum hirsutum [J]. Biotechnology Bulletin, 2024, 40(7): 259-272. |
| [2] | HUANG Dan, JIANG Shan, PENG Tao. Cloning of FfCYP98 Gene and Its Functional Analysis in Folioceros fuciformis [J]. Biotechnology Bulletin, 2024, 40(7): 273-284. |
| [3] | WANG Yu-shu, ZHAO Lin-lin, ZHAO Shuang, HU Qi, BAI Hui-xia, WANG Huan, CAO Ye-ping, FAN Zhen-yu. Cloning and Expression Analysis of BrCYP83B1 Gene in Chinese Cabbage [J]. Biotechnology Bulletin, 2024, 40(6): 152-160. |
| [4] | TIAN Sheng-ni, ZHANG Qin, DONG Yu-fei, DING Zhou, YE Ai-hua, ZHANG Ming-zhu. Effects of Acid Mine Drainage on Physicochemical Factors and Nitrogen-fixing Microorganisms in the Root Zone of Mature Rice [J]. Biotechnology Bulletin, 2024, 40(6): 271-280. |
| [5] | HAO Si-yi, ZHANG Jun-ke, WANG Bin, QU Peng-yan, LI Rui-de, CHENG Chun-zhen. Cloning and Expression Analysis of Banana EARLY FLOWERING 3(ELF3)Genes [J]. Biotechnology Bulletin, 2024, 40(5): 131-140. |
| [6] | KONG De-ting, QI Xiao-han, LIU Xing-lei, LI Li-ping, HU Feng-yi, HUANG Li-yu, QIN Shi-wen. Comparison and Analysis of Endophytic Bacterial Communities in Different Perennial Rice Varieties [J]. Biotechnology Bulletin, 2024, 40(5): 225-236. |
| [7] | DU Ze-guang, REN Shao-wen, ZHANG Feng-qin, LI Mei-lan, LI Gai-zhen, QI Xian-hui. Cloning,Expression and Functional Identification of BrMLP328 Gene in Brassica rapa subsp. pekinensis [J]. Biotechnology Bulletin, 2024, 40(4): 122-129. |
| [8] | LIU Huan-huan, YANG Li-chun, LI Huo-gen. Cloning and Functional Analysis of LtMYB305 in Liriodendron tulipifera [J]. Biotechnology Bulletin, 2024, 40(4): 179-188. |
| [9] | ZHONG Yun, LIN Chun, LIU Zheng-jie, DONG Chen-wen-hua, MAO Zi-chao, LI Xing-yu. Cloning and Prokaryotic Expression Analysis of Asparagus Saponin Synthesis Related Glycosyltransferase Genes [J]. Biotechnology Bulletin, 2024, 40(4): 255-263. |
| [10] | YANG Qi, WEI Zi-di, SONG Juan, TONG Kun, YANG Liu, WANG Jia-han, LIU Hai-yan, LUAN Wei-jiang, MA Xuan. Construction and Transcriptomic Analysis of Rice Histone H1 Triple Mutant [J]. Biotechnology Bulletin, 2024, 40(4): 85-96. |
| [11] | LI Xing-rong, TAN Zhi-bing, ZHAO Yan, LI Yao-kui, ZHAO Bing-ran, TANG Li. Cloning and Functional Analysis of OsLCT3, a Low-affinity Cation Transporter Gene of Rice [J]. Biotechnology Bulletin, 2024, 40(4): 97-109. |
| [12] | LIU Jia-ning, LI Meng, YANG Xin-sen, WU Wei, PEI Xin-wu, YUAN Qian-hua. Impact of Different Water Management Cultivation Methods on the Rhizosphere Bacteria Community of Shanlan Upland Rice [J]. Biotechnology Bulletin, 2024, 40(3): 242-250. |
| [13] | LI Xue, LI Rong-ou, KONG Mei-yi, HUANG Lei. The Growth Promoting Effect of Bacillus amyloliquefaciens SQ-2 on Rice [J]. Biotechnology Bulletin, 2024, 40(2): 109-119. |
| [14] | YANG Yan, HU Yang, LIU Ni-ru, YIN Lu, YANG Rui, WANG Peng-fei, MU Xiao-peng, ZHANG Shuai, CHENG Chun-zhen, ZHANG Jian-cheng. Cloning and Functional Analysis of MbbZIP43 Gene in ‘Hongmantang’ Red-flesh Apple [J]. Biotechnology Bulletin, 2024, 40(2): 146-159. |
| [15] | ZHANG Chao, WANG Zi-rui, SUN Ya-li, MAO Xin-chen, TANG Jia-qi, YU Heng-xiu. Functional Study of Vitamin B1 Synthesis-related Gene OsTHIC in Rice [J]. Biotechnology Bulletin, 2024, 40(2): 99-108. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||