








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (10): 191-197.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0673
Previous Articles Next Articles
JIAO Dan-rong1,2( ), MA Meng-xue2,3, HE Bai-shui2, XIE Long2, ZUO Er-wei2(
), MA Meng-xue2,3, HE Bai-shui2, XIE Long2, ZUO Er-wei2( )
)
Received:2024-07-15
Online:2024-10-26
Published:2024-11-20
Contact:
ZUO Er-wei
E-mail:m15834071899@163.com;zuoerwei@caas.cn
JIAO Dan-rong, MA Meng-xue, HE Bai-shui, XIE Long, ZUO Er-wei. Editing Features of High-fidelity CRISPR/Cas9 System in Poultry Cells[J]. Biotechnology Bulletin, 2024, 40(10): 191-197.
| 引物名称Primer name | sgRNA打靶位点sgRNA target-sites(5'-3') | 引物序列Primer sequence(5'-3') | 基因Gene |
|---|---|---|---|
| SgRNA1 | ATAACTGCCGTTCAGATGAT | F:ATAACTGCCGTTCAGATGAT R:ATCATCTGAACGGCAGTTATC | ANP32B |
| SgRNA2 | GGAGATTGGAGATGGACAGC | F:GAGATTGGAGATGGACAGC R:GCTGTCCATCTCCAATCTCC | ANP32B |
| SgRNA3 | GTCCATCCCCATCTGCCTCA | F:TCCATCCCCATCTGCCTCA R:TGAGGCAGATGGGGATGGAC | ANP32B |
| SgRNA4 | GGAAGAGGAGGATGATGATG | F:GAAGAGGAGGATGATGATG R:CATCATCATCCTCCTCTTCC | ANP32B |
| SgRNA5 | TGATCTGGAAGGTGAAGAGG | F:TGATCTGGAAGGTGAAGAGG R:CCTCTTCACCTTCCAGATCAC | ANP32B |
| SgRNA6 | TGAACAATATGGTACTGTGA | F:TGAACAATATGGTACTGTGA R:TCACAGTACCATATTGTTCAC | DAZL |
Table 1 Primer list
| 引物名称Primer name | sgRNA打靶位点sgRNA target-sites(5'-3') | 引物序列Primer sequence(5'-3') | 基因Gene |
|---|---|---|---|
| SgRNA1 | ATAACTGCCGTTCAGATGAT | F:ATAACTGCCGTTCAGATGAT R:ATCATCTGAACGGCAGTTATC | ANP32B |
| SgRNA2 | GGAGATTGGAGATGGACAGC | F:GAGATTGGAGATGGACAGC R:GCTGTCCATCTCCAATCTCC | ANP32B |
| SgRNA3 | GTCCATCCCCATCTGCCTCA | F:TCCATCCCCATCTGCCTCA R:TGAGGCAGATGGGGATGGAC | ANP32B |
| SgRNA4 | GGAAGAGGAGGATGATGATG | F:GAAGAGGAGGATGATGATG R:CATCATCATCCTCCTCTTCC | ANP32B |
| SgRNA5 | TGATCTGGAAGGTGAAGAGG | F:TGATCTGGAAGGTGAAGAGG R:CCTCTTCACCTTCCAGATCAC | ANP32B |
| SgRNA6 | TGAACAATATGGTACTGTGA | F:TGAACAATATGGTACTGTGA R:TCACAGTACCATATTGTTCAC | DAZL |

Fig. 1 Construction process of plasmid sgRNA and mapping of different high-fidelity editors a: Construction process of sgRNA plasmid; b: the structure of plasmid editors
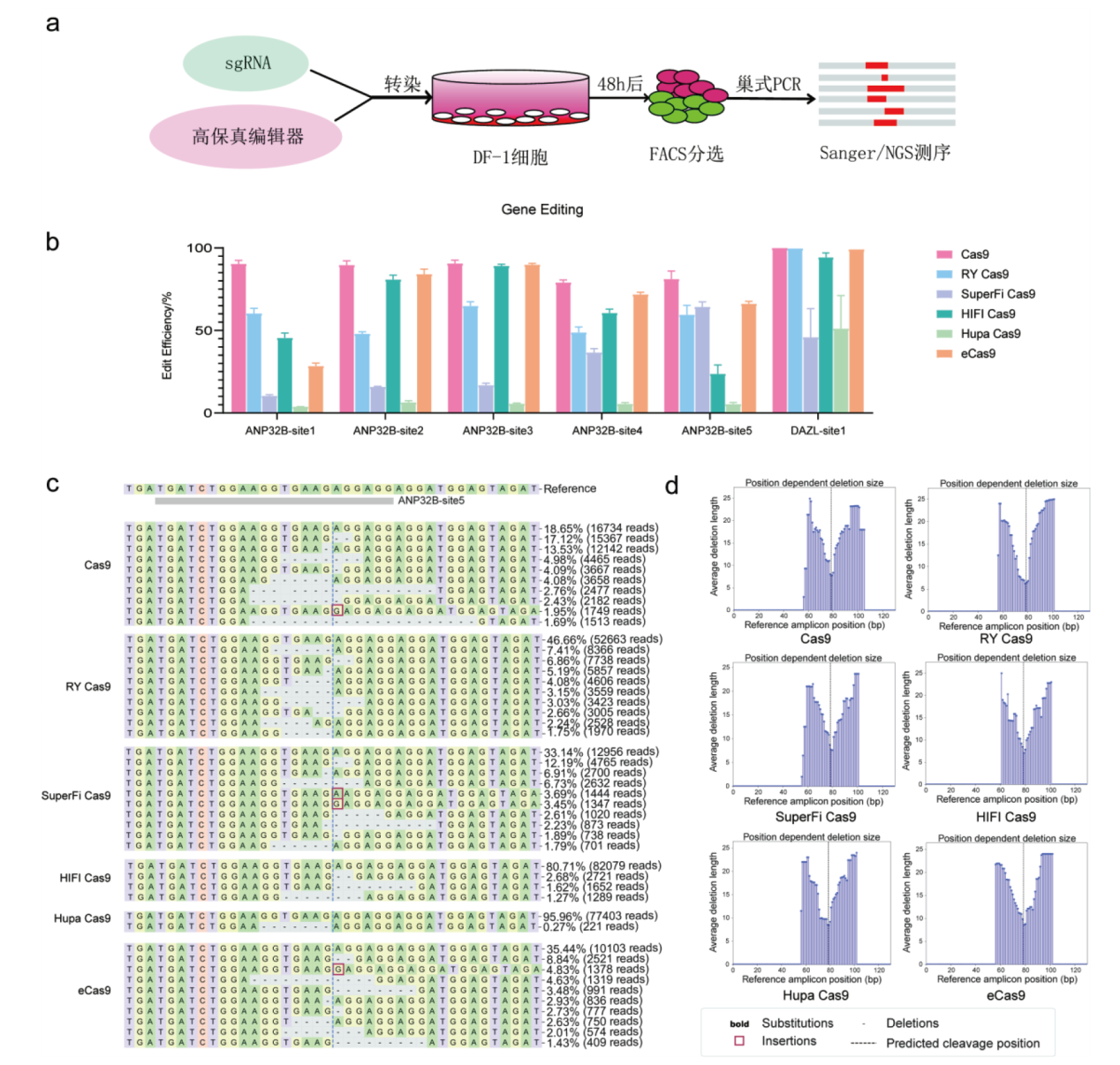
Fig. 2 Editing features of different high-fidelity editors in chicken gene ANP32B and DAZL a: The experiment schedule of gene editing in chicken cells for high-fidelity editors; b: editing efficiency comparing of different high-fidelity editors in different target sites from ANP32B and DAZL; c: detailed editing characteristics of different high-fidelity editors in ANP32B-site5 target site; d: the editing windows of different high-fidelity editors in ANP32B-site5
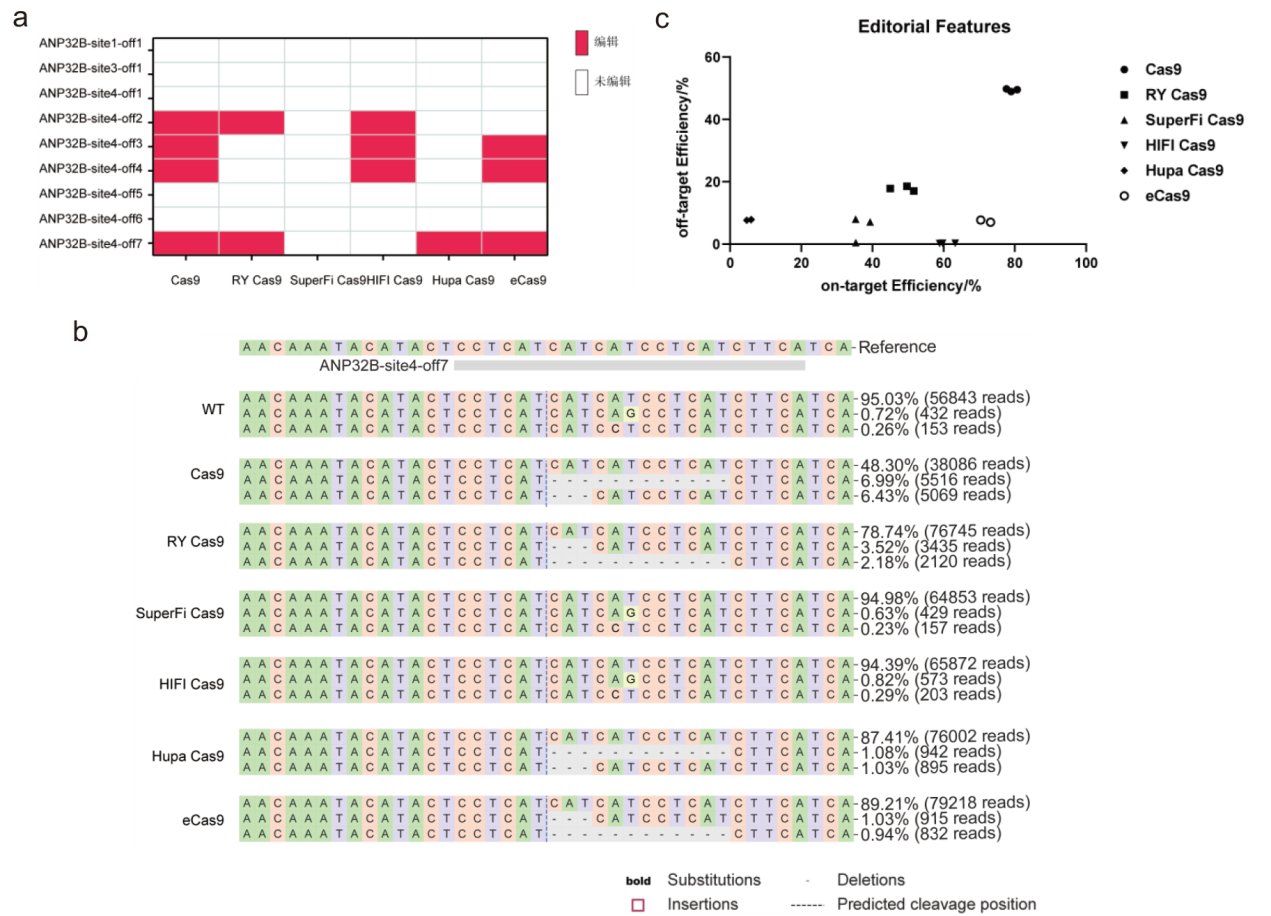
Fig. 3 Off-target editing of different high-fidelity editors in editing chicken's ANP32B and DAZL loci a: The off-target effects at ANP32B-site1, 3, 4 of different Cas9 variants; b: the detail off-target characteristics induced by different Cas9 varaints at ANP32B-site4-off7 locus; c: joint analysis of the on-target and off-target effects of Cas9 variants
| [1] | 邓蓉. 我国家禽产业现状分析及未来发展趋势[J]. 现代化农业, 2020(3): 68-71. |
| Deng R. Present situation analysis and future development trend of poultry industry in China[J]. Mod Agric, 2020(3): 68-71. | |
| [2] | 刘旗. 我国畜禽遗传资源面临的问题及解决途径[J]. 农业技术与装备, 2012(15): 43-45. |
| Liu Q. Problems and solutions of livestock and poultry genetic resources in China[J]. Agric Technol Equip, 2012(15): 43-45. | |
| [3] |
Zhang F. CRISPR-Cas9: prospects and challenges[J]. Hum Gene Ther, 2015, 26(7): 409-410.
doi: 10.1089/hum.2015.29002.fzh pmid: 26176430 |
| [4] |
Fu YF, Foden JA, Khayter C, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells[J]. Nat Biotechnol, 2013, 31(9): 822-826.
doi: 10.1038/nbt.2623 pmid: 23792628 |
| [5] |
Anderson KR, Haeussler M, Watanabe C, et al. CRISPR off-target analysis in genetically engineered rats and mice[J]. Nat Methods, 2018, 15(7): 512-514.
doi: 10.1038/s41592-018-0011-5 pmid: 29786090 |
| [6] | Zhang BY, Wang CY, Zhang Y, et al. A CRISPR-engineered swine model of COL2A1 deficiency recapitulates altered early skeletal developmental defects in humans[J]. Bone, 2020, 137: 115450. |
| [7] | Tang HH, Wang DQ, Shu YL. Structural insights into Cas9 mismatch: promising for development of high-fidelity Cas9 variants[J]. Sig Transduct Target Ther, 2022, 7: 271. |
| [8] |
Vakulskas CA, Dever DP, Rettig GR, et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells[J]. Nat Med, 2018, 24(8): 1216-1224.
doi: 10.1038/s41591-018-0137-0 pmid: 30082871 |
| [9] |
Ikeda A, Fujii W, Sugiura K, et al. High-fidelity endonuclease variant HypaCas9 facilitates accurate allele-specific gene modification in mouse zygotes[J]. Commun Biol, 2019, 2: 371.
doi: 10.1038/s42003-019-0627-8 pmid: 31633062 |
| [10] |
Walton RT, Christie KA, Whittaker MN, et al. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants[J]. Science, 2020, 368(6488): 290-296.
doi: 10.1126/science.aba8853 pmid: 32217751 |
| [11] | Bravo JPK, Liu MS, Hibshman GN, et al. Structural basis for mismatch surveillance by CRISPR-Cas9[J]. Nature, 2022, 603(7900): 343-347. |
| [12] |
Zong Y, Wang YP, Li C, et al. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion[J]. Nat Biotechnol, 2017, 35(5): 438-440.
doi: 10.1038/nbt.3811 pmid: 28244994 |
| [13] |
Tang X, Ren QR, Yang LJ, et al. Single transcript unit CRISPR 2.0 systems for robust Cas9 and Cas12a mediated plant genome editing[J]. Plant Biotechnol J, 2019, 17(7): 1431-1445.
doi: 10.1111/pbi.13068 pmid: 30582653 |
| [14] | Kim N, Choi S, Kim S, et al. Deep learning models to predict the editing efficiencies and outcomes of diverse base editors[J]. Nat Biotechnol, 2024, 42(3): 484-497. |
| [15] |
Idoko-Akoh A, Goldhill DH, Sheppard CM, et al. Creating resistance to avian influenza infection through genome editing of the ANP32 gene family[J]. Nat Commun, 2023, 14(1): 6136.
doi: 10.1038/s41467-023-41476-3 pmid: 37816720 |
| [16] |
Rengaraj D, Zheng YH, Kang KS, et al. Conserved expression pattern of chicken DAZL in primordial germ cells and germ-line cells[J]. Theriogenology, 2010, 74(5): 765-776.
doi: 10.1016/j.theriogenology.2010.04.001 pmid: 20537692 |
| [17] |
Kulcsár PI, Tálas A, Huszár K, et al. Crossing enhanced and high fidelity SpCas9 nucleases to optimize specificity and cleavage[J]. Genome Biol, 2017, 18(1): 190.
doi: 10.1186/s13059-017-1318-8 pmid: 28985763 |
| [18] | Hu JH, Miller SM, Geurts MH, et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity[J]. Nature, 2018, 556(7699): 57-63. |
| [19] | Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease[J]. Nature, 2014, 513(7519): 569-573. |
| [20] |
Zhang DB, Zhang HW, Li TD, et al. Perfectly matched 20-nucleotide guide RNA sequences enable robust genome editing using high-fidelity SpCas9 nucleases[J]. Genome Biol, 2017, 18(1): 191.
doi: 10.1186/s13059-017-1325-9 pmid: 29020979 |
| [21] | 戴小芳. 鸡病流行特点及防治措施分析[J]. 新农民, 2024(11):126-128. |
| Dai XF. Analysis of epidemic characteristics and preventive measures of chicken diseases[J]. New Farmer, 2024(11):126-128. | |
| [22] | 张迎冰, 张成图, 吴英, 等. 单碱基编辑技术的原理、发展及其在家畜育种中的应用[J]. 生物工程学报, 2023, 39(1):19-33. |
| Zhang YB, Zhang CT, Wu Y, et al. Principles and development of single base editing technology and its application in livestock breeding[J]. Journal of Biological Engineering, 2023, 39(1):19-33. | |
| [23] | Chen JS, Dagdas YS, Kleinstiver BP, et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy[J]. Nature, 2017, 550(7676): 407-410. |
| [24] | Kim N, Kim HK, Lee S, et al. Prediction of the sequence-specific cleavage activity of Cas9 variants[J]. Nat Biotechnol, 2020, 38(11): 1328-1336. |
| [25] |
Kulcsár PI, Tálas A, Ligeti Z, et al. SuperFi-Cas9 exhibits remarkable fidelity but severely reduced activity yet works effectively with ABE8e[J]. Nat Commun, 2022, 13(1): 6858.
doi: 10.1038/s41467-022-34527-8 pmid: 36369279 |
| [1] | ZHOU Ai-ting, PENG Rui-qi, WANG Fang, WU Jian-rong, MA Huan-cheng. Analysis of Metabolic Differences of Biocontrol Strain DZY6715 at Different Growth Stages [J]. Biotechnology Bulletin, 2023, 39(9): 225-235. |
| [2] | CHEN Xiao-ling, LIAO Dong-qing, HUANG Shang-fei, CHEN Ying, LU Zhi-long, CHEN Dong. Advances in CRISPR/Cas9 System Modifying Saccharomycescerevisiae [J]. Biotechnology Bulletin, 2023, 39(8): 148-158. |
| [3] | YANG Lan, ZHANG Chen-xi, FAN Xue-wei, WANG Yang-guang, WANG Chun-xiu, LI Wen-ting. Gene Cloning, Expression Pattern, and Promoter Activity Analysis of Chicken BMP15 [J]. Biotechnology Bulletin, 2023, 39(4): 304-312. |
| [4] | CHEN Chu-wen, LI Jie, ZHAO Rui-peng, LIU Yuan, WU Jin-bo, LI Zhi-xiong. Cloning, Tissue Expression Profile and Function Prediction of GPX3 Gene in Tibetan Chicken [J]. Biotechnology Bulletin, 2023, 39(3): 311-320. |
| [5] | DU Zhen-wei, ZHU Shuai-peng, MA Xiang-fei, LI Dong-hua, SUN Gui-rong. Cloning,Expression and Bioinformatics Analysis of the CDS Region of Chicken CEBPA Gene [J]. Biotechnology Bulletin, 2021, 37(8): 203-212. |
| [6] | ZHANG Chen, ZUO Qi-sheng, ZOU Yi-chen, ZHAO Juan-juan, ZHANG Ya-ni, LI Bi-chun. Study on the Function of Glycolysis in Inducing Chicken PGCLC in vitro Formation [J]. Biotechnology Bulletin, 2021, 37(6): 163-170. |
| [7] | YIN Jun-lei, ZHANG Yan-fang, ZOU Fan-yu, PAN Peng-tao, DUAN Yan-hong, QIU Shu-xing. Construction and Immunoprotection of sptP Deletion Mutant of Salmonella Pullorum [J]. Biotechnology Bulletin, 2021, 37(2): 122-128. |
| [8] | ZHANG Chen, LEI Zhan, LI Kai, SHANG Ying, XU Wen-tao. Research Progress on Off-target Effects and Detection Techniques in CRISPR/Cas9 Systems [J]. Biotechnology Bulletin, 2020, 36(3): 78-87. |
| [9] | WANG Jing, DAI Dong, WU Shu-geng, ZHANG Hai-jun, QI Guang-hai. Advances in Successional Development and Early Establishment of the Chicken Intestinal Microbiota [J]. Biotechnology Bulletin, 2020, 36(2): 1-8. |
| [10] | YU Hai-liang, ZOU Wen-bin, WANG Xiao-hui, LIN Yu-xin, DAI Guo-jun, ZHANG Tao, ZHANG Gen-xi, XIE Kai-zhou, WANG Jin-yu, SHI Hui-qiang. RNA Sequencing Analysis of Cecum Tissues of Jinghai Yellow Chickens Infected by E. tenella [J]. Biotechnology Bulletin, 2019, 35(11): 64-71. |
| [11] | ZHANG Ya-nan, LIN Ya-qiu, XU Qing, XU Ya-ou, HE Qing-hua. Correlation Analysis Between IRX3 Gene Expression and Intramuscular Fat Deposition in Tibetan Chicken [J]. Biotechnology Bulletin, 2018, 34(9): 219-223. |
| [12] | WU Yan, HAO Ya-qiao, WEI Xuan, SHEN Qi, LIU Ye-fei, WANG Sheng-hou, ZHAO Hong-xin. The Advantages and Limitations of CRISPR/Cas9-based Gene Editing Technology [J]. Biotechnology Bulletin, 2018, 34(5): 1-8. |
| [13] | BI Yan-zhen, XIAO Hong-wei, ZHANG Li-ping, REN Hong-yan, HUA Zai-dong, HUA Wen-jun, WANG Zheng, NIU Min-jie, LIN Zheng-yun, REN Xi-dong, SUN Li-hua, ZHENG Xin-min. Applications and Challenges of Gene Editing in Non-human Primate Models [J]. Biotechnology Bulletin, 2018, 34(5): 48-56. |
| [14] | DONG Wei-peng, WANG Jun-shi, ZHANG Shao-hua, YAN Jiong. Research Progress of CRISPR System and Its Application in Mice [J]. Biotechnology Bulletin, 2018, 34(5): 57-63. |
| [15] | LI Dong-hua, WANG Xin-lei ,LI Zhuan-jian ,SUN Gui-rong ,KANG Xiang-tao, YAN Feng-bin. Research Advances on Whole Genome Sequencing of Chicken [J]. Biotechnology Bulletin, 2017, 33(7): 35-39. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||