








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (12): 267-279.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0438
Previous Articles Next Articles
WANG Ruo-Ruo1,2( ), QU Peng-Kun1, ZHANG Xin1, WANG Luo1, ZHU Ying1, TIAN Shuang-Yi3, ZHAO De-Gang2,4(
), QU Peng-Kun1, ZHANG Xin1, WANG Luo1, ZHU Ying1, TIAN Shuang-Yi3, ZHAO De-Gang2,4( )
)
Received:2025-04-24
Online:2025-12-26
Published:2026-01-06
Contact:
ZHAO De-Gang
E-mail:ruozhu_w@163.com;dgzhao@gzu.edu.cn
WANG Ruo-Ruo, QU Peng-Kun, ZHANG Xin, WANG Luo, ZHU Ying, TIAN Shuang-Yi, ZHAO De-Gang. Molecular Characterization, Expression Profiling, and Protein Interaction Analysis of the EuGIF1 Gene in Eucommia ulmoides[J]. Biotechnology Bulletin, 2025, 41(12): 267-279.
引物名称 Primer name | 序列 Sequence (5′‒3′) |
|---|---|
| cdsEuGIF1-F | ctagaggatctcgaggcATGCAGCAGCACCTGATG |
| cdsEuGIF1-R | cgtctgtacacctaggTCAGTTTCCATCATCAGAA |
| EuGIF1gfp-F | ttggagaggacacgcATGCAGCAGCACCTGATG |
| EuGIF1gfp-R | cttctcccttacccatGTTTCCATCATCAGAAGCCT |
| YN-GIF1-F | gcatttaaatctcgaggATGCAGCAGCACCTGATG |
| YN-GIF1-R | gtggcgatggatcttctGTTTCCATCATCAGAAGC |
| YC-GRF5-F | gaggaggacctgctttATGAACGTGAACGCGATGA |
| YC-GRF5-R | acgccggacgggtaccgCCAGTAGGGTCTGGAGT |
| AD-GIF1-F | ggccatggaggccagtATGCAGCAGCACCTGATG |
| AD-GIF1-R | gcagctcgagctcgatGTTTCCATCATCAGAAGCC |
| BD-GRF5-F | tatggccatggaggccATGAACGTGAACGCGATGA |
| BD-GRF5-R | gccgctgcaggtcgacCCAGTAGGGTCTGGAGTT |
| YS22-GFP-R | aggaagctttccagtagtgca |
| termi-R | aagaccggcaacaggattc |
| cdsEuGIF1-F2 | CAGCTGGGAATGAGCTCTGG |
| EuGIF1-qRT-F | CAGCAGTATTCGGCACTACAG |
| EuGIF1-qRT--R | CCTCCACTCCCACCCATT |
| EuGIF2-qRT--F | CCATCACCAGCAGCATCAAT |
| EuGIF2-qRT--R | CACCGCCACCTCCGATAT |
| EuGIF3-qRT--F | ACAATGGCTCAGCAACAACA |
| EuGIF3-qRT--R | GTGGTGGAAGTGGAGTAGGT |
| EuACTIN-qRT--F | GTGTTATGGTTGGGATGGG |
| EuACTIN-qRT--R | TGCTGACTATGCCGTGTTC |
Table 1 Table of primer sequences
引物名称 Primer name | 序列 Sequence (5′‒3′) |
|---|---|
| cdsEuGIF1-F | ctagaggatctcgaggcATGCAGCAGCACCTGATG |
| cdsEuGIF1-R | cgtctgtacacctaggTCAGTTTCCATCATCAGAA |
| EuGIF1gfp-F | ttggagaggacacgcATGCAGCAGCACCTGATG |
| EuGIF1gfp-R | cttctcccttacccatGTTTCCATCATCAGAAGCCT |
| YN-GIF1-F | gcatttaaatctcgaggATGCAGCAGCACCTGATG |
| YN-GIF1-R | gtggcgatggatcttctGTTTCCATCATCAGAAGC |
| YC-GRF5-F | gaggaggacctgctttATGAACGTGAACGCGATGA |
| YC-GRF5-R | acgccggacgggtaccgCCAGTAGGGTCTGGAGT |
| AD-GIF1-F | ggccatggaggccagtATGCAGCAGCACCTGATG |
| AD-GIF1-R | gcagctcgagctcgatGTTTCCATCATCAGAAGCC |
| BD-GRF5-F | tatggccatggaggccATGAACGTGAACGCGATGA |
| BD-GRF5-R | gccgctgcaggtcgacCCAGTAGGGTCTGGAGTT |
| YS22-GFP-R | aggaagctttccagtagtgca |
| termi-R | aagaccggcaacaggattc |
| cdsEuGIF1-F2 | CAGCTGGGAATGAGCTCTGG |
| EuGIF1-qRT-F | CAGCAGTATTCGGCACTACAG |
| EuGIF1-qRT--R | CCTCCACTCCCACCCATT |
| EuGIF2-qRT--F | CCATCACCAGCAGCATCAAT |
| EuGIF2-qRT--R | CACCGCCACCTCCGATAT |
| EuGIF3-qRT--F | ACAATGGCTCAGCAACAACA |
| EuGIF3-qRT--R | GTGGTGGAAGTGGAGTAGGT |
| EuACTIN-qRT--F | GTGTTATGGTTGGGATGGG |
| EuACTIN-qRT--R | TGCTGACTATGCCGTGTTC |
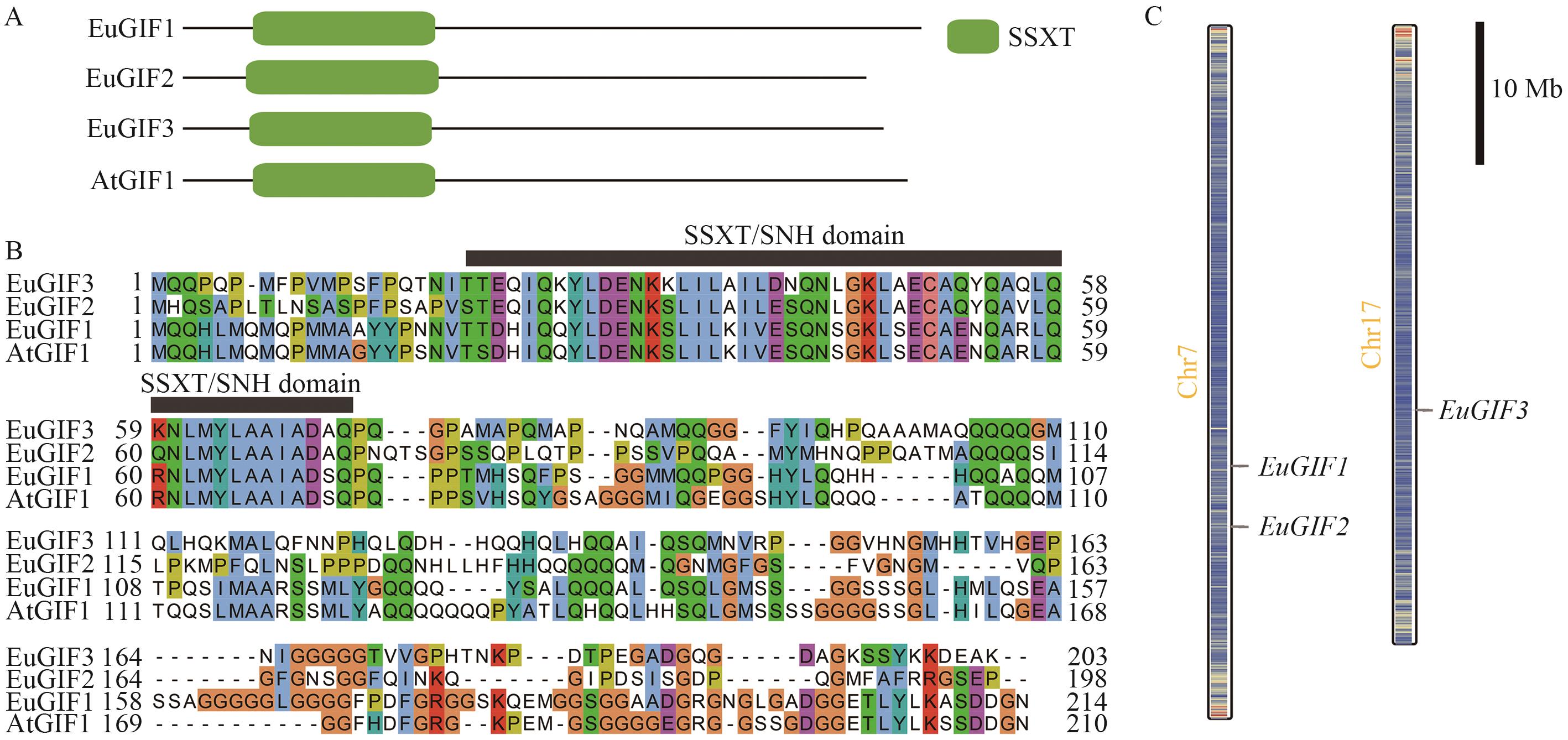
Fig. 1 Distribution of conserved domains in the EuGIFs protein sequences and chromosomal distribution of the genes in E. ulmoidesA: Distribution of conserved domains in the EuGIFs protein sequences. B: Amino acid sequence alignment of EuGIFs proteins. C: Chromosomal distribution of the EuGIFs genes
基因 名称 Gene name | 基因ID Gene ID | 氨基酸长度 Number of amino acids (aa) | 蛋白分子量 Molecular weight (Da) | 等电点 Theoretical pI | 不稳定指数 Instability index | 脂肪族氨基酸指数 Aliphatic index | 疏水性平均值 Grand average of hydropathicity | 亚细胞定位 Subcellular location |
|---|---|---|---|---|---|---|---|---|
| EuGIF1 | evm.model.Chr7.1262 | 203 | 22 275.03 | 6.62 | 61.24 | 59.26 | -0.833 | Nucleus |
| EuGIF2 | evm.model.Chr7.1483 | 198 | 21 579.25 | 6.01 | 83.45 | 64.14 | -0.565 | Nucleus |
| EuGIF3 | evm.model.Chr17.1343 | 214 | 22 651.04 | 5.84 | 71.27 | 54.86 | -0.678 | Nucleus |
Table 2 Physicochemical properties of the EuGIFs proteins in E. ulmoides
基因 名称 Gene name | 基因ID Gene ID | 氨基酸长度 Number of amino acids (aa) | 蛋白分子量 Molecular weight (Da) | 等电点 Theoretical pI | 不稳定指数 Instability index | 脂肪族氨基酸指数 Aliphatic index | 疏水性平均值 Grand average of hydropathicity | 亚细胞定位 Subcellular location |
|---|---|---|---|---|---|---|---|---|
| EuGIF1 | evm.model.Chr7.1262 | 203 | 22 275.03 | 6.62 | 61.24 | 59.26 | -0.833 | Nucleus |
| EuGIF2 | evm.model.Chr7.1483 | 198 | 21 579.25 | 6.01 | 83.45 | 64.14 | -0.565 | Nucleus |
| EuGIF3 | evm.model.Chr17.1343 | 214 | 22 651.04 | 5.84 | 71.27 | 54.86 | -0.678 | Nucleus |
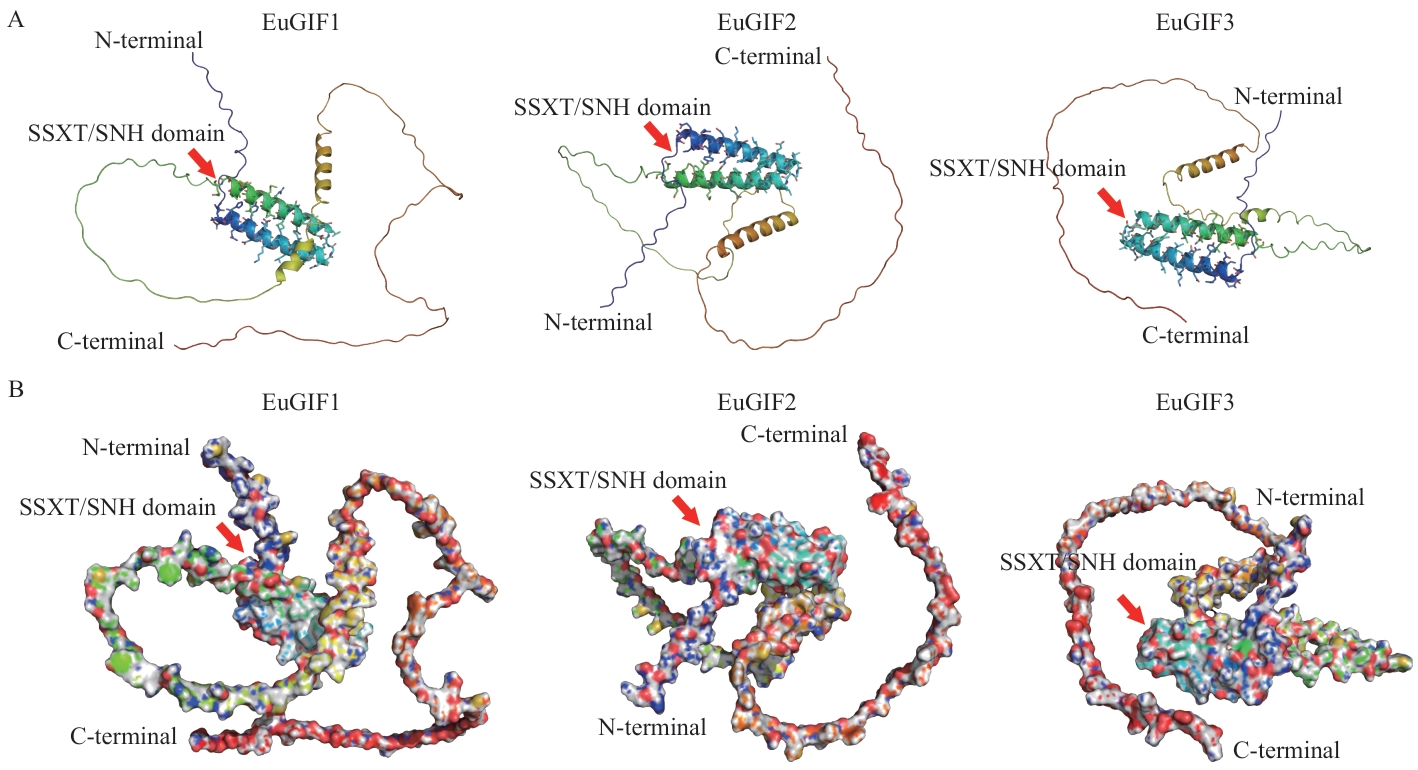
Fig. 2 Three-dimensional spatial structure diagram of the EuGIFs proteinA: Cartoon representation of the 3D structure of EuGIFs protein. Regions corresponding to the SSXT/SNH domains are indicated by arrows. B: Surface representation of the 3D structure of EuGIFs protein. The arrows denote the amino acids with pLDDT (predicted local distance difference test) values>90, which correspond to the amino acids depicted as stick structures in Fig. A
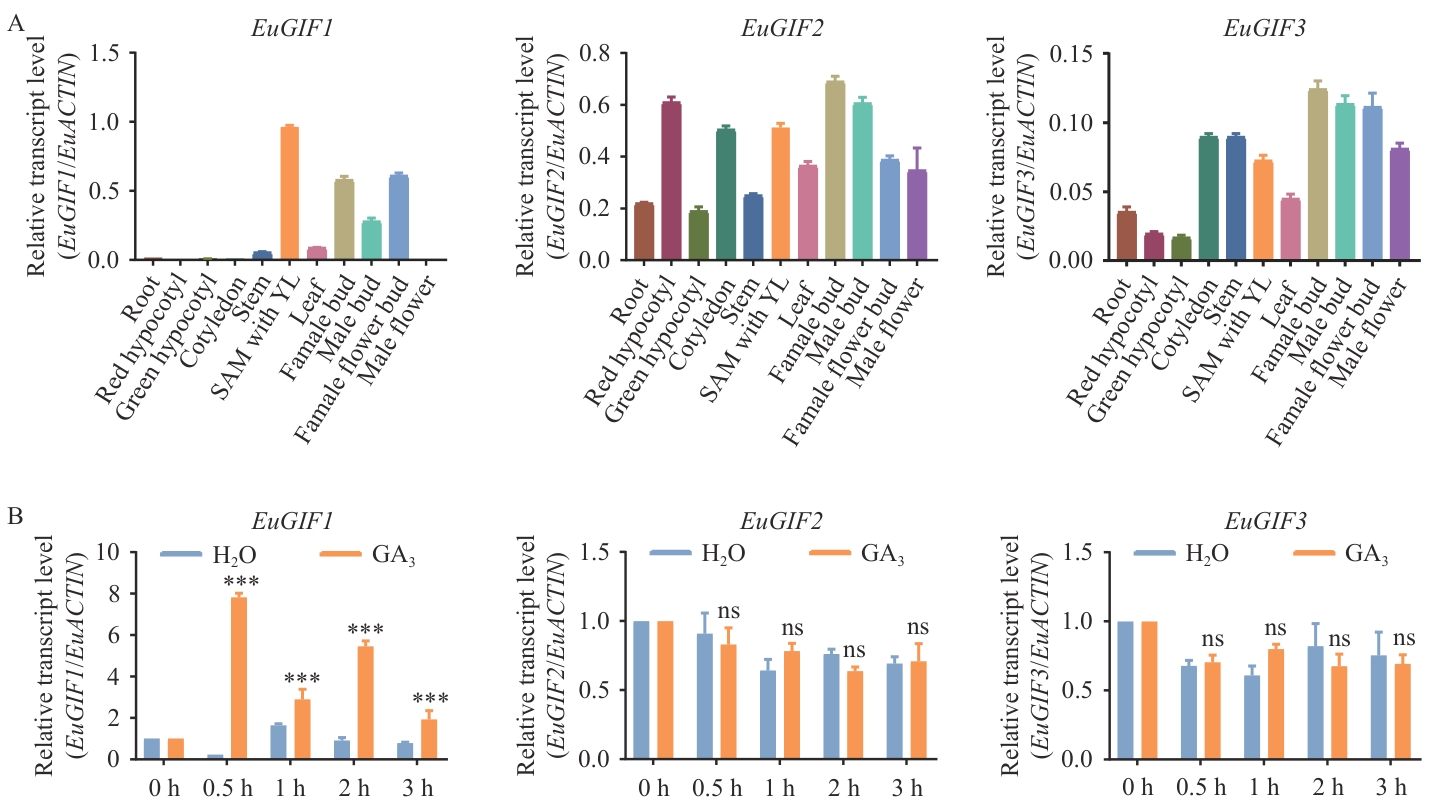
Fig. 4 Expression pattern of EuGIFs genes in E. ulmoidesA: Expression profiles of the EuGIFs gene in different tissues of E. ulmoides. B: Expression profiles of the EuGIFs gene in young leaves of seedlings following exogenous GA3 treatment. *** P<0.001, ns indicates no statistically significant difference (P>0.05)
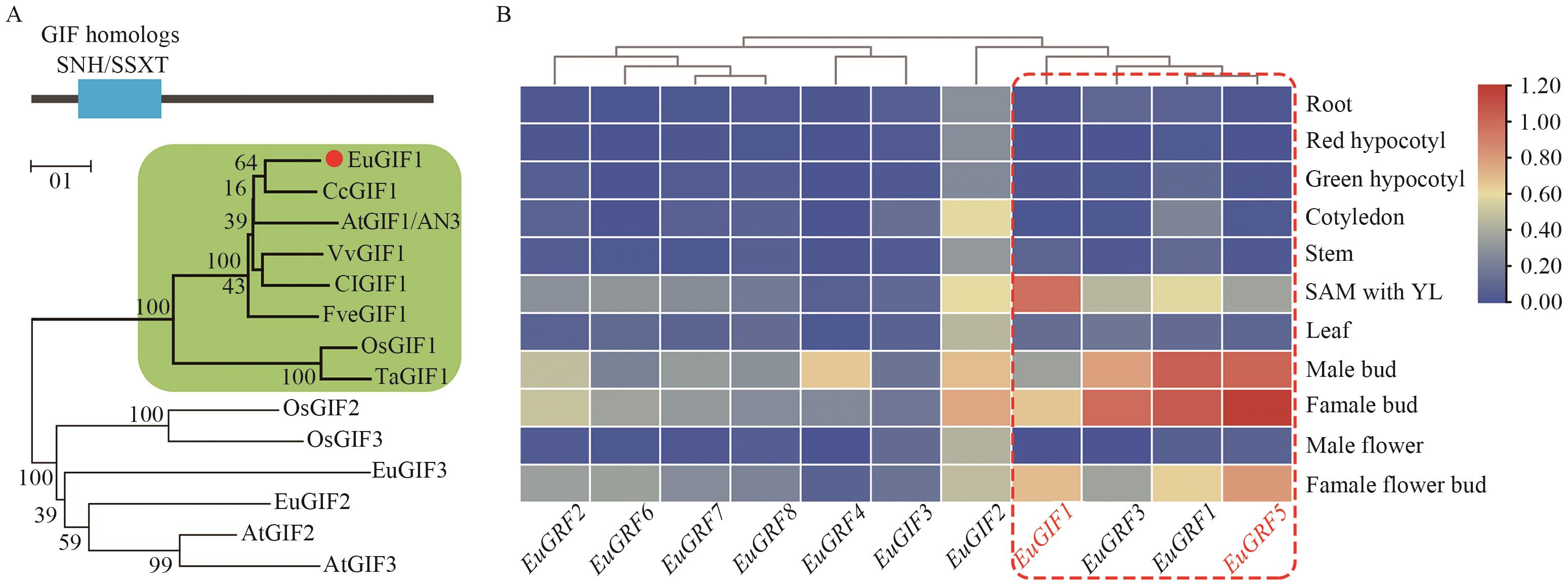
Fig. 5 Phylogenetic analysis of GIF homologs and gene co-expression profiles of EuGIF and EuCRFsA: Neighbor-joining phylogenetic tree constructed using full-length protein sequences, with all proteins containing the conserved SNH/SSXT domain. The green-shaded region highlights AtGIF1 homologs, and EuGIF1 is marked in red. The species abbreviations in the protein names, Eu, Cc, At, Vv, Cl, Fve, Os, and Ta, correspond to Eucommia ulmoides, Citrus clementina, Arabidopsis thaliana, Vitis vinifera, Citrullus lanatus, Fragaria vesca, Oryza sativa, and Triticum aestivum, respectively. B: Expression patterns of EuGIFs and EuGRFs, with the red dashed box indicating genes having high and correlated expression in meristematic tissues. The color gradient (blue-yellow-red) indicate expression levels (low-medium-high) of EuGRFs
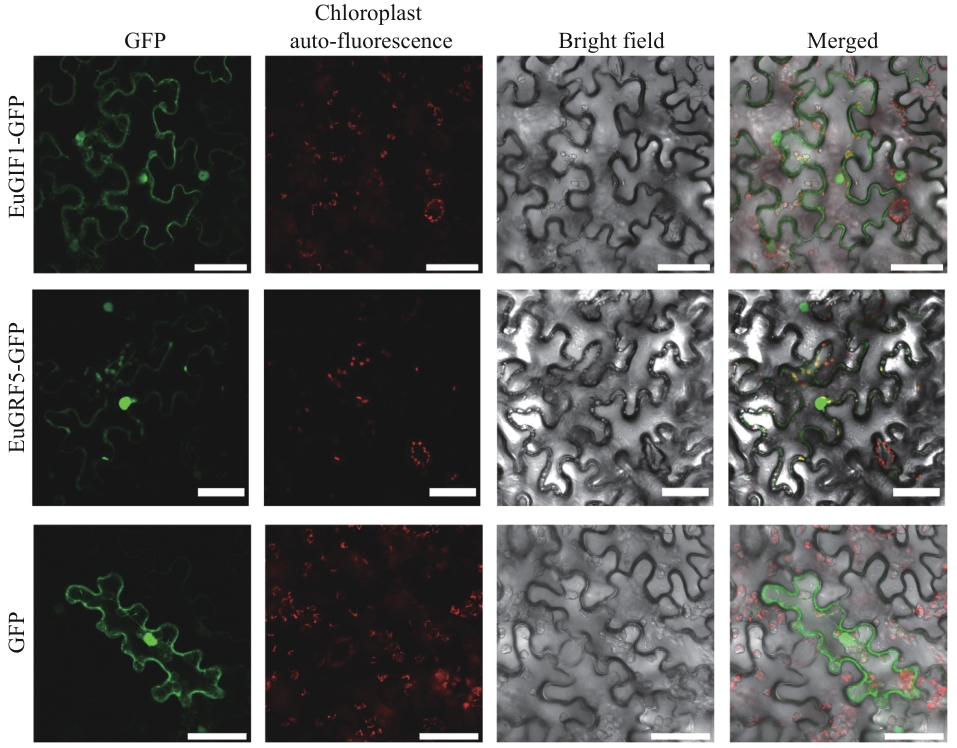
Fig. 6 Subcellular localization of GFP-tagged proteins in the epidermal cells of tobacco leafThe figure displays four imaging modalities: GFP fluorescence (green, excitation/emission: 488/510 nm); chloroplast autofluorescence (red, excitation/emission: 640/675 nm); bright field (grayscale) and merged image. Scale bar: 50 μm
蛋白模型编号 Number | 结构偏差 RMSD (Å) | 亲和力 Score |
|---|---|---|
| 1 | 50.52 | -809.33 |
| 2 | 31.22 | -774.96 |
| 3 | 69.01 | -764.03 |
| 4 | 60.97 | -758.89 |
| 5 | 55.71 | -755.58 |
| 6 | 79.41 | -749.46 |
| 7 | 64.33 | -747.77 |
| 8 | 62.85 | -735.99 |
| 9 | 62.97 | -732.66 |
| 10 | 68.86 | -732.20 |
Table 3 Structural analysis of EuGIF1-EuGRF5 protein-protein interaction predicted by HDOCK
蛋白模型编号 Number | 结构偏差 RMSD (Å) | 亲和力 Score |
|---|---|---|
| 1 | 50.52 | -809.33 |
| 2 | 31.22 | -774.96 |
| 3 | 69.01 | -764.03 |
| 4 | 60.97 | -758.89 |
| 5 | 55.71 | -755.58 |
| 6 | 79.41 | -749.46 |
| 7 | 64.33 | -747.77 |
| 8 | 62.85 | -735.99 |
| 9 | 62.97 | -732.66 |
| 10 | 68.86 | -732.20 |
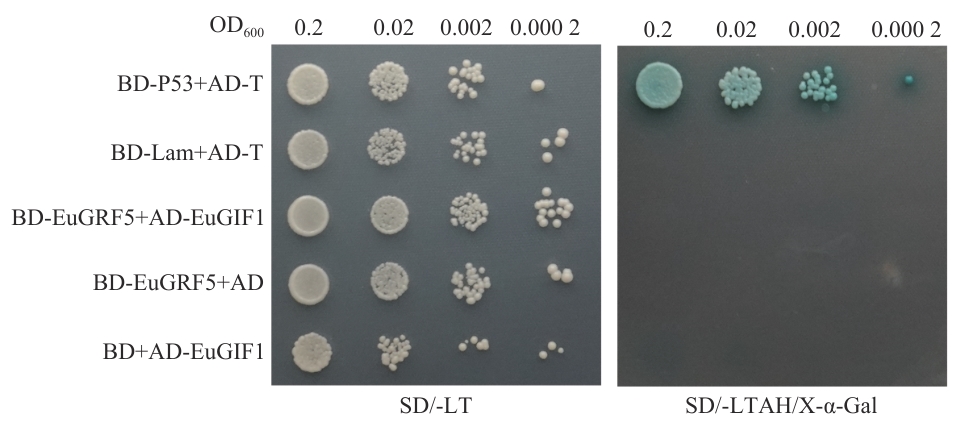
Fig. 7 Yeast two-hybrid assay for protein-protein interaction between EuGIF1 and EuGRF5SD/-LT: Synthetic dropout medium deficient in leucine (-L) and tryptophan (-T). SD/-LTAH/X-α-Gal: Stringent selection medium with quadruple dropout (-L/-T/-A/-H) supplemented with 5-bromo-4-chloro-3-indolyl-α-D-galactopyranoside (X-α-Gal). The indicated OD600 values (0.2, 0.02, 0.002, and 0.000 2) represent 10-fold serial dilutions of yeast cultures prior to spotting assay. Positive control: BD-P53+AD-T; negative control 1: BD-Lam+AD-T; test group: BD-EuGRF5+AD-EuGIF1; negative control 2: BD-EuGRF5+AD/BD+AD-EuGIF1
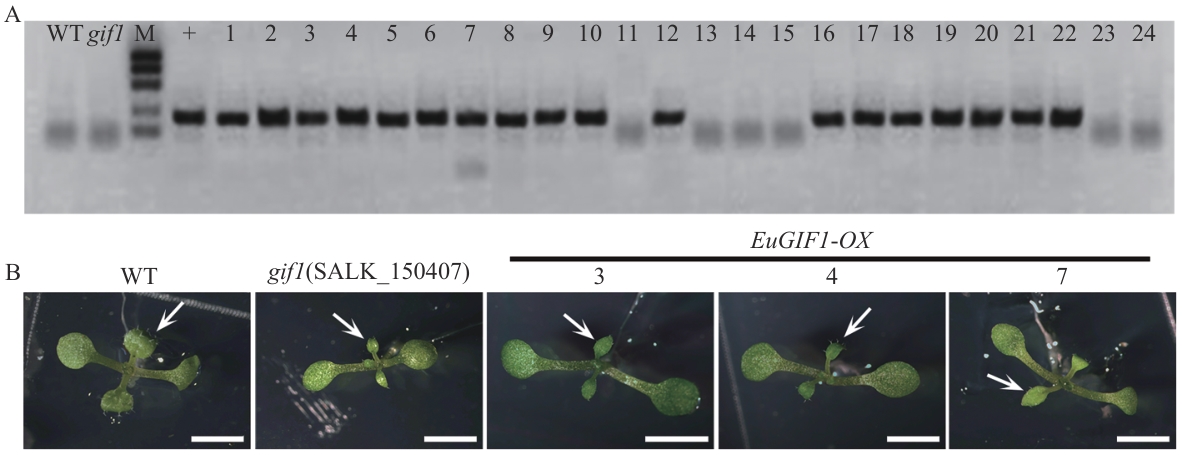
Fig. 8 Genetic complementation of the A. thalianagif1 mutant by EuGIF1A: Genotyping results of T3 generation transgenic plants for the complementation test. B: Phenotypic characterization of 12 d T3 transgenic seedlings grown in medium, with arrows indicating the wild type, mutant, and three representative T3 homozygous positive transgenic lines for juvenile leaves. Scale bar: 0.5 cm
| [1] | Horiguchi G, Kim GT, Tsukaya H. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana [J]. Plant J, 2005, 43(1): 68-78. |
| [2] | Omidbakhshfard MA, Proost S, Fujikura U, et al. Growth-regulating factors (GRFs): a small transcription factor family with important functions in plant biology [J]. Mol Plant, 2015, 8(7): 998-1010. |
| [3] | Rodriguez RE, Ercoli MF, Debernardi JM, et al. Growth-regulating factors, a transcription factor family regulating more than just plant growth [M]// Plant Transcription Factors. Amsterdam: Elsevier, 2016: 269-280. |
| [4] | Debernardi JM, Tricoli DM, Ercoli MF, et al. A GRF-GIF chimeric protein improves the regeneration efficiency of transgenic plants [J]. Nat Biotechnol, 2020, 38(11): 1274-1279. |
| [5] | Feng Q, Xiao L, He YZ, et al. Highly efficient, genotype-independent transformation and gene editing in watermelon (Citrullus lanatus) using a chimeric ClGRF4-GIF1 gene [J]. J Integr Plant Biol, 2021, 63(12): 2038-2042. |
| [6] | Pan WB, Cheng ZT, Han ZG, et al. Efficient genetic transformation and CRISPR/Cas9-mediated genome editing of watermelon assisted by genes encoding developmental regulators [J]. J Zhejiang Univ Sci B, 2022, 23(4): 339-344. |
| [7] | 郭苏平, 田美萍, 丁静. GRF-GIF嵌合体在草莓遗传转化中的应用 [J/OL]. 分子植物育种, 2022. . |
| Guo SP, Tian MP, Ding J. Application of GRF-GIF Chimera in genetic transformation of strawberry [J/OL]. Mol Plant Breed, 2022. . | |
| [8] | Bull T, Debernardi J, Reeves M, et al. GRF-GIF chimeric proteins enhance in vitro regeneration and Agrobacterium-mediated transformation efficiencies of lettuce (Lactuca spp.) [J]. Plant Cell Rep, 2023, 42(3): 629-643. |
| [9] | Zhao Y, Cheng P, Liu Y, et al. A highly efficient soybean transformation system using GRF3-GIF1 chimeric protein [J]. J Integr Plant Biol, 2025, 67(1): 3-6. |
| [10] | Lee BH, Ko JH, Lee SM, et al. The Arabidopsis GRF-INTERACTING FACTOR gene family performs an overlapping function in determining organ size as well as multiple developmental properties [J]. Plant Physiol, 2009, 151(2): 655-668. |
| [11] | Kim JH, Kende H. A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis [J]. Proc Natl Acad Sci USA, 2004, 101(36): 13374-13379. |
| [12] | Horiguchi G, Nakayama H, Ishikawa N, et al. ANGUSTIFOLIA3 plays roles in adaxial/abaxial patterning and growth in leaf morphogenesis [J]. Plant Cell Physiol, 2011, 52(1): 112-124. |
| [13] | Kanei MR, Horiguchi G, Tsukaya H. Stable establishment of Cotyledon identity during embryogenesis in Arabidopsis by ANGUSTIFOLIA3 and HANABA TARANU [J]. Development, 2012, 139(13): 2436-2446. |
| [14] | Lee BH, Wynn AN, Franks RG, et al. The Arabidopsis thaliana GRF-INTERACTING FACTOR gene family plays an essential role in control of male and female reproductive development [J]. Dev Biol, 2014, 386(1): 12-24. |
| [15] | Lee SJ, Lee BH, Jung JH, et al. GROWTH-REGULATINGFACTOR and GRF-INTERACTING FACTOR specify meristematic cells of gynoecia and anthers [J]. Plant Physiol, 2018, 176(1): 717-729. |
| [16] | Liu HH, Xiong F, Duan CY, et al. Importin β4 mediates nuclear import of GRF-interacting factors to control ovule development in Arabidopsis [J]. Plant Physiol, 2019, 179(3): 1080-1092. |
| [17] | Liu ZP, Li N, Zhang YY, et al. Transcriptional repression of GIF1 by the KIX-PPD-MYC repressor complex controls seed size in Arabidopsis [J]. Nat Commun, 2020, 11(1): 1846. |
| [18] | Li SC, Gao FY, Xie KL, et al. The OsmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in rice [J]. Plant Biotechnol J, 2016, 14(11): 2134-2146. |
| [19] | He ZS, Zeng J, Ren Y, et al. OsGIF1 positively regulates the sizes of stems, leaves, and grains in rice [J]. Front Plant Sci, 2017, 8: 1730. |
| [20] | Shimano S, Hibara KI, Furuya T, et al. Conserved functional control, but distinct regulation, of cell proliferation in rice and Arabidopsis leaves revealed by comparative analysis of GRF-INTERACTING FACTOR 1 orthologs [J]. Development, 2018, 145(7): dev159624. |
| [21] | Zhang D, Sun W, Singh R, et al. GRF-interacting factor1 regulates shoot architecture and meristem determinacy in maize [J]. Plant Cell, 2018, 30(2): 360-374. |
| [22] | 崔迪. 玉米gif1突变体雄性不育的分子机理研究 [D]. 武汉: 华中农业大学, 2021. |
| Cui D. A study on molecular mechanism of male sterility in maize gif1 mutant [D]. Wuhan: Huazhong Agricultural University, 2021. | |
| [23] | 郑媛媛. 玉米GIF1基因调控雌花序形态建成和小花发育的分子机理研究 [D]. 武汉: 华中农业大学, 2019. |
| Zheng YY. A study on molecular mechanism of GIF1 in the regulation of female inflorescence architecture and floral development in maize [D]. Wuhan: Huazhong Agricultural University, 2019. | |
| [24] | Li MF, Zheng YY, Cui D, et al. GIF1 controls ear inflorescence architecture and floral development by regulating key genes in hormone biosynthesis and meristem determinacy in maize [J]. BMC Plant Biol, 2022, 22(1): 127. |
| [25] | 张立全, 张浩林, 李丛丛, 等. 谷子生长调节因子互作因子基因家族生物信息学分析 [J]. 福建农业学报, 2021, 36(8): 878-883. |
| Zhang LQ, Zhang HL, Li CC, et al. Bioinformatics of growth-interacting factor genes in foxtail millet [J]. Fujian J Agric Sci, 2021, 36(8): 878-883. | |
| [26] | 邱姗, 王文新, 江梅, 等. 杉木GRF和GIF基因家族鉴定、表达与互作分析 [J]. 核农学报, 2024, 38(7): 1224-1233. |
| Qiu S, Wang WX, Jiang M, et al. Identification, expression and interaction analysis of GRF and GIF gene families in Cunninghamia lanceolata [J]. J Nucl Agric Sci, 2024, 38(7): 1224-1233. | |
| [27] | 吴洁, 淮东欣, 薛晓梦, 等. 花生GRF和GIF基因家族的鉴定与表达分析 [J/OL]. 分子植物育种, 2023. . |
| Wu J, Huai DX, Xue XM, et al. Genome-wide identification and expression analysis of GRF and GIF gene families in peanut [J/OL]. Mol Plant Breed, 2023. . | |
| [28] | 杨宇琦. 胡杨GIF基因家族鉴定及PeGIF3对叶形调控的研究 [D]. 阿拉尔: 塔里木大学, 2024. |
| Yang YQ. Identification of GIF gene family and regulation of leaf shape by PeGIF3 in Populus euphratica [D]. Ala'er: Tarim University, 2024. | |
| [29] | 杜仲产业研究课题组. 杜仲生物产业发展现状与前景 [J]. 中国林业产业, 2022(11): 18-47. |
| Research Group of Eucommia ulmoides Industry. Present situation and prospect of Eucommia ulmoides biological industry development [J]. China For Ind, 2022(11): 18-47. | |
| [30] | Wuyun TN, Wang L, Liu HM, et al. The hardy rubber tree genome provides insights into the evolution of polyisoprene biosynthesis [J]. Mol Plant, 2018, 11(3): 429-442. |
| [31] | Li Y, Wei HR, Yang J, et al. High-quality de novo assembly of the Eucommia ulmoides haploid genome provides new insights into evolution and rubber biosynthesis [J]. Hortic Res, 2020, 7(1): 183. |
| [32] | Du QX, Wu ZX, Liu PF, et al. The chromosome-level genome of Eucommia ulmoides provides insights into sex differentiation and α-linolenic acid biosynthesis [J]. Front Plant Sci, 2023, 14: 1118363. |
| [33] | 王珞, 朱英, 赵德刚. 杜仲CRISPR/Cas9基因编辑系统的建立 [J]. 植物生理学报, 2023, 59(8): 1566-1574. |
| Wang L, Zhu Y, Zhao DG. An efficient CRISPR/Cas9 mutagenesis system for Eucommia ulmoides [J]. Plant Physiol J, 2023, 59(8): 1566-1574. | |
| [34] | Jumper J, Evans R, Pritzel A, et al. Highly accurate protein structure prediction with AlphaFold [J]. Nature, 2021, 596(7873): 583-589. |
| [35] | Evans R, O'Neill M, Pritzel A, et al. Protein complex prediction with AlphaFold-Multimer [J]. BioRxiv, 2022. DOI: https://doi.org/10.1101/2021.10.04.463034 . |
| [36] | Yan YM, Tao HY, He JH, et al. The HDOCK server for integrated protein-protein docking [J]. Nat Protoc, 2020, 15(5): 1829-1852. |
| [37] | Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data [J]. Mol Plant, 2020, 13(8): 1194-1202. |
| [38] | Wang RR, Lu N, Liu CG, et al. MtGSTF7, a TT19-like GST gene, is essential for accumulation of anthocyanins, but not proanthocyanins in Medicago truncatula [J]. J Exp Bot, 2022, 73(12): 4129-4146. |
| [39] | Zhou SL, Yang TQ, Mao YW, et al. The F-box protein MIO1/SLB1 regulates organ size and leaf movement in Medicago truncatula [J]. J Exp Bot, 2021, 72(8): 2995-3011. |
| [40] | Qin L, Chen HF, Wu QF, et al. Identification and exploration of the GRF and GIF families in maize and foxtail millet [J]. Physiol Mol Biol Plants, 2022, 28(9): 1717-1735. |
| [41] | Wu WQ, Li J, Wang Q, et al. Growth-regulating factor 5 (GRF5)-mediated gene regulatory network promotes leaf growth and expansion in poplar [J]. New Phytol, 2021, 230(2): 612-628. |
| [42] | Wang RR, Zhu Y, Zhao DG. Genome-wide identification and expression analysis of growth-regulating factors in Eucommia ulmoides oliver (du-Zhong) [J]. Plants, 2024, 13(9): 1185. |
| [43] | 何文广, 苏印泉, 徐咏梅, 等. 外源激素影响杜仲叶中次生代谢物含量的研究 [J]. 西北林学院学报, 2009, 24(6): 121-123. |
| He WG, Su YQ, Xu YM, et al. Effects of external hormones on secondary metabolites in the leaf of Eucommia ulmoides [J]. J Northwest For Univ, 2009, 24(6): 121-123. | |
| [44] | 徐咏梅. 外源激素对杜仲次生代谢物含量的影响 [D]. 杨凌: 西北农林科技大学, 2007. |
| Xu YM. The effect of external hormones on the content of secondary metabolites in Eucommia ulmoides Oliv [D]. Yangling: Northwest A & F University, 2007. | |
| [45] | 张强, 苏印泉, 徐咏梅. 采样时间对赤霉素处理杜仲叶次生代谢物含量的影响 [J]. 西北林学院学报, 2010, 25(6): 130-133. |
| Zhang Q, Su YQ, Xu YM. Effect of harvesting time on contents of secondary metabolites in the leaf of Eucommia ulmoides treated by gibberellin acid [J]. J Northwest For Univ, 2010, 25(6): 130-133. | |
| [46] | 魏艳秀, 刘慧东, 杜红岩, 等. 外源激素提高杜仲叶中有效成分含量的效果 [J]. 中南林业科技大学学报, 2017, 37(5): 42-49. |
| Wei YX, Liu HD, Du HY, et al. Efficacy of exogenous hormones enhancing active ingredients in Eucommia ulmoides leaves [J]. J Cent South Univ For Technol, 2017, 37(5): 42-49. |
| [1] | WANG Fang, SHAO Hui-ru, LYU Lin-long, ZHAO Dian, HU Zhen, LYU Jian-zhen, JIANG Liang. Establishment of TurboID Proximity Labeling Technology in Plants and Bacteria [J]. Biotechnology Bulletin, 2025, 41(9): 44-53. |
| [2] | CHENG Ting-ting, LIU Jun, WANG Li-li, LIAN Cong-long, WEI Wen-jun, GUO Hui, WU Yao-lin, YANG Jing-fan, LAN Jin-xu, CHEN Sui-qing. Genome-wide Identification of the Chalcone Isomerase Gene Family in Eucommia ulmoides and Analysis of Their Expression Patterns [J]. Biotechnology Bulletin, 2025, 41(9): 242-255. |
| [3] | WANG Cong-huan, WU Guo-qiang, WEI Ming. Functional Mechanism of Plant CBL in Regulating the Responses to Abiotic and Biotic Stresses [J]. Biotechnology Bulletin, 2025, 41(7): 1-16. |
| [4] | HUANG Dan-dan, WU Yun-yi, ZOU Jian-hua, YU Ting, ZHU Yan-hui, YANG Mei-hong, DONG Wen-li, GAO Dong-li. Cloning and Interaction Analysis of StPTST2a Gene in Potato [J]. Biotechnology Bulletin, 2025, 41(7): 172-180. |
| [5] | QIN Yue, YANG Yan, ZHANG Lei, LU Li-li, LI Xian-ping, JIANG Wei. Identification and Comparative Analysis of the StGAox Genes in Diploid and Tetraploid Potatoes [J]. Biotechnology Bulletin, 2025, 41(3): 146-160. |
| [6] | DUAN Ruo-xin, CHEN Ying-ying, LIN Jin-xing, LI Rui-li. Advance on Seed Dormancy and Its Regulation Mechanism [J]. Biotechnology Bulletin, 2025, 41(12): 16-26. |
| [7] | WANG Ya-ping, JIN Lan, HAO Jin-feng, CHANG Ming, WANG Yan-dan, GAO Feng. Identification and Expression Characteristics Analysis of CmRGLG Gene Family in Melon [J]. Biotechnology Bulletin, 2025, 41(12): 156-167. |
| [8] | YANG Wei, ZHAO Li-fen, TANG Bing, ZHOU Lin-bi, YANG Juan, MO Chuan-yuan, ZHANG Bao-hui, LI Fei, RUAN Song-lin, DENG Ying. Genome-wide Identification and Expression Analysis of the SRO Gene Family in Brassica juncea L. [J]. Biotechnology Bulletin, 2024, 40(8): 129-141. |
| [9] | WU Ding-jie, CHEN Ying-ying, XU Jing, LIU Yuan, ZHANG Hang, LI Rui-li. Research Progress in Plant Gibberellin Oxidase and Its Functions [J]. Biotechnology Bulletin, 2024, 40(7): 43-54. |
| [10] | JIN Yun-qian, WANG Bin, GUO Shu-lei, ZHAO Lin-xi, HAN Zan-ping. Research Progress in Gibberellin Regulation on Maize Seed Vigor [J]. Biotechnology Bulletin, 2023, 39(1): 84-94. |
| [11] | LI Yi-dan, SHAN Xiao-hui. Gibberellin Metabolism Regulation and Green Revolution [J]. Biotechnology Bulletin, 2022, 38(2): 195-204. |
| [12] | LI Meng, CHEN Yue, HU Feng-rong. Research Progress in miR159-GAMYB Regulating Plants Growth and Development [J]. Biotechnology Bulletin, 2021, 37(9): 234-247. |
| [13] | JIA Jian-lei, CHEN Qian, JIN Ji-peng, YUAN Zan, ZHANG Li-ping. Eukaryotic Expression of BMPR1B in Sheep and Identification of Its Interaction Proteins [J]. Biotechnology Bulletin, 2019, 35(12): 94-104. |
| [14] | GAO Xiu-hua, FU Xiang-dong. Research Progress for the Gibberellin Signaling and Action on Plant Growth and Development [J]. Biotechnology Bulletin, 2018, 34(7): 1-13. |
| [15] | HE Yi-xue, LIU Wen, SHEN Xiang-ling. Effects of GA and B9 on the Growth and Development of Potato Tubers [J]. Biotechnology Bulletin, 2018, 34(7): 66-73. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||