








Biotechnology Bulletin ›› 2026, Vol. 42 ›› Issue (1): 218-229.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0726
Previous Articles Next Articles
LI Jian-bin1,2,3( ), HOU Jia-e1,2, LI Lei-lin1,2, AI Ming-tao1,2, LIU Tian-tai1,2, CUI Xiu-ming1,2,3(
), HOU Jia-e1,2, LI Lei-lin1,2, AI Ming-tao1,2, LIU Tian-tai1,2, CUI Xiu-ming1,2,3( ), YANG Qian1,2(
), YANG Qian1,2( )
)
Received:2025-07-07
Online:2026-01-26
Published:2026-02-04
Contact:
CUI Xiu-ming, YANG Qian
E-mail:3274327386@qq.com;sanqi37@vip.sina.com;qian1226@vip.sina.com
LI Jian-bin, HOU Jia-e, LI Lei-lin, AI Ming-tao, LIU Tian-tai, CUI Xiu-ming, YANG Qian. Genome-wide Identification of Panax notoginseng Lipoxygenases Coupled in Response to Methyl-jasmonate and Wounding[J]. Biotechnology Bulletin, 2026, 42(1): 218-229.
基因名 Gene name | 正向引物 Forward primer (5′-3′) | 反向引物 Reverse primer (5′-3′) | 产物大小 Product (bp) |
|---|---|---|---|
| PnACTIN2 | TCCAAGGGTGAATATGATGAATCG | AACCTCTCCAAAGAGAATTTCTGAGT | 199 |
| PnLOX1 | TAGAGGGCCTTCCTGCTGAT | GCTTGGAGCTCCCTGTCATT | 208 |
| PnLOX2 | CCCAATGTGGTCAGCAGGAT | TGGGCCATATATCTCCGGGT | 207 |
| PnLOX3 | GAGGGATGGCAGTTGAGGAC | CATGGCCCTTTTCTCGGACT | 210 |
| PnLOX4 | CACTCCAGCTGAAGAGGGTG | GCGGAAGTGAGGTTGGAGAA | 199 |
| PnLOX5 | CCTTTATCTCCGGCGATGCT | AACCTTGGTGGCCTTATCGG | 158 |
| PnLOX6 | ACCGACAACTAGGTGCGATG | CTGCTGGCAATCCTTCCTGA | 210 |
| PnLOX7 | CCTTGACCCTGTGAGCTACG | GTTTTCCTTTTCCCGGTGGC | 269 |
| PnLOX8 | GCAGGTGGAAGCGTTGAAAG | AACTTCAGGCCATAGGGTGC | 167 |
| PnLOX9 | AAGCAGGGGTCAGTAACAGC | AGATCCCCTGACCAATCCCA | 244 |
| PnLOX10 | TCGCCTCCATGAGTTTCCAC | AGGTAGAGGAGTGTGCGACT | 234 |
Table 1 Primer sequences for RT-qPCR
基因名 Gene name | 正向引物 Forward primer (5′-3′) | 反向引物 Reverse primer (5′-3′) | 产物大小 Product (bp) |
|---|---|---|---|
| PnACTIN2 | TCCAAGGGTGAATATGATGAATCG | AACCTCTCCAAAGAGAATTTCTGAGT | 199 |
| PnLOX1 | TAGAGGGCCTTCCTGCTGAT | GCTTGGAGCTCCCTGTCATT | 208 |
| PnLOX2 | CCCAATGTGGTCAGCAGGAT | TGGGCCATATATCTCCGGGT | 207 |
| PnLOX3 | GAGGGATGGCAGTTGAGGAC | CATGGCCCTTTTCTCGGACT | 210 |
| PnLOX4 | CACTCCAGCTGAAGAGGGTG | GCGGAAGTGAGGTTGGAGAA | 199 |
| PnLOX5 | CCTTTATCTCCGGCGATGCT | AACCTTGGTGGCCTTATCGG | 158 |
| PnLOX6 | ACCGACAACTAGGTGCGATG | CTGCTGGCAATCCTTCCTGA | 210 |
| PnLOX7 | CCTTGACCCTGTGAGCTACG | GTTTTCCTTTTCCCGGTGGC | 269 |
| PnLOX8 | GCAGGTGGAAGCGTTGAAAG | AACTTCAGGCCATAGGGTGC | 167 |
| PnLOX9 | AAGCAGGGGTCAGTAACAGC | AGATCCCCTGACCAATCCCA | 244 |
| PnLOX10 | TCGCCTCCATGAGTTTCCAC | AGGTAGAGGAGTGTGCGACT | 234 |
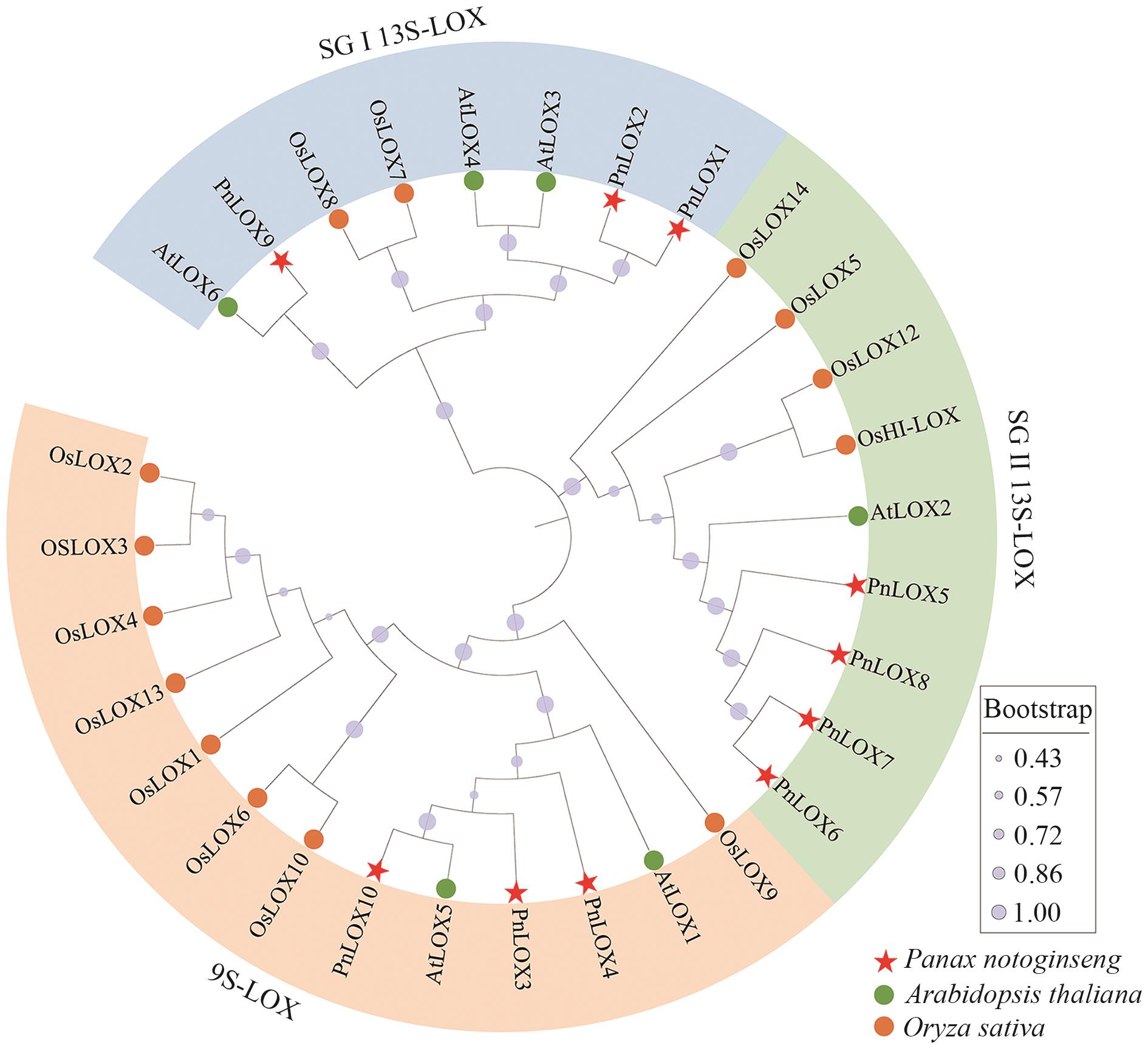
Fig. 1 Phylogenetic tree of LOX proteins of P. notoginseng, A. thaliana, and Oryza sativaDifferent species are indicated in different shapes and colors. The various colors indicate the different subgroups
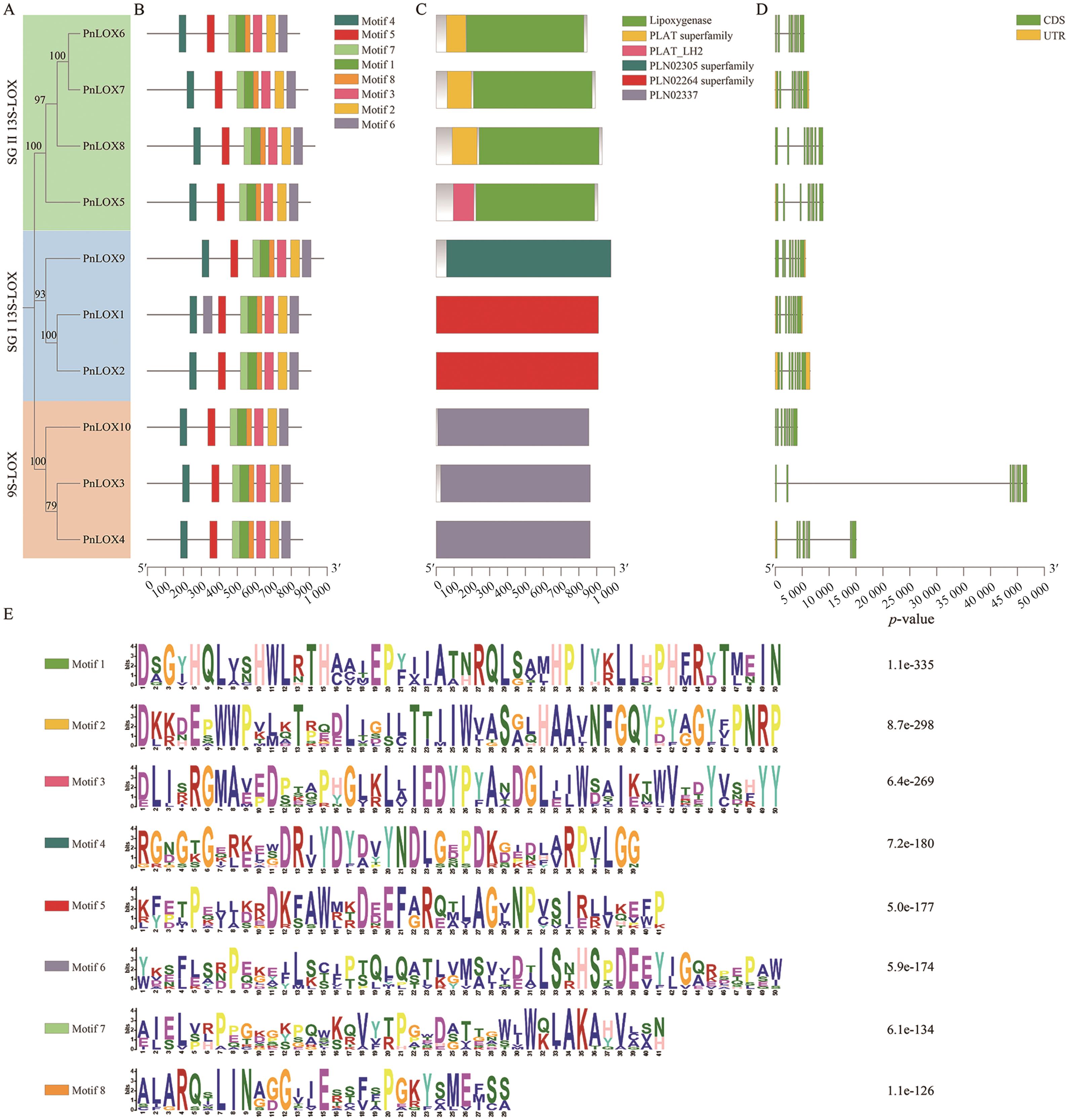
Fig. 2 Phylogenetic relationship, conserved motifs, conserved domain and gene structure of the PnLOXsA: Phylogenetic tree of PnLOXs. B: Conserved motifs of PnLOXs. C: Conserved domain of PnLOXs. D: Gene structure of PnLOXs. D: Amino acid sequence of PnLOXs conserved motifs
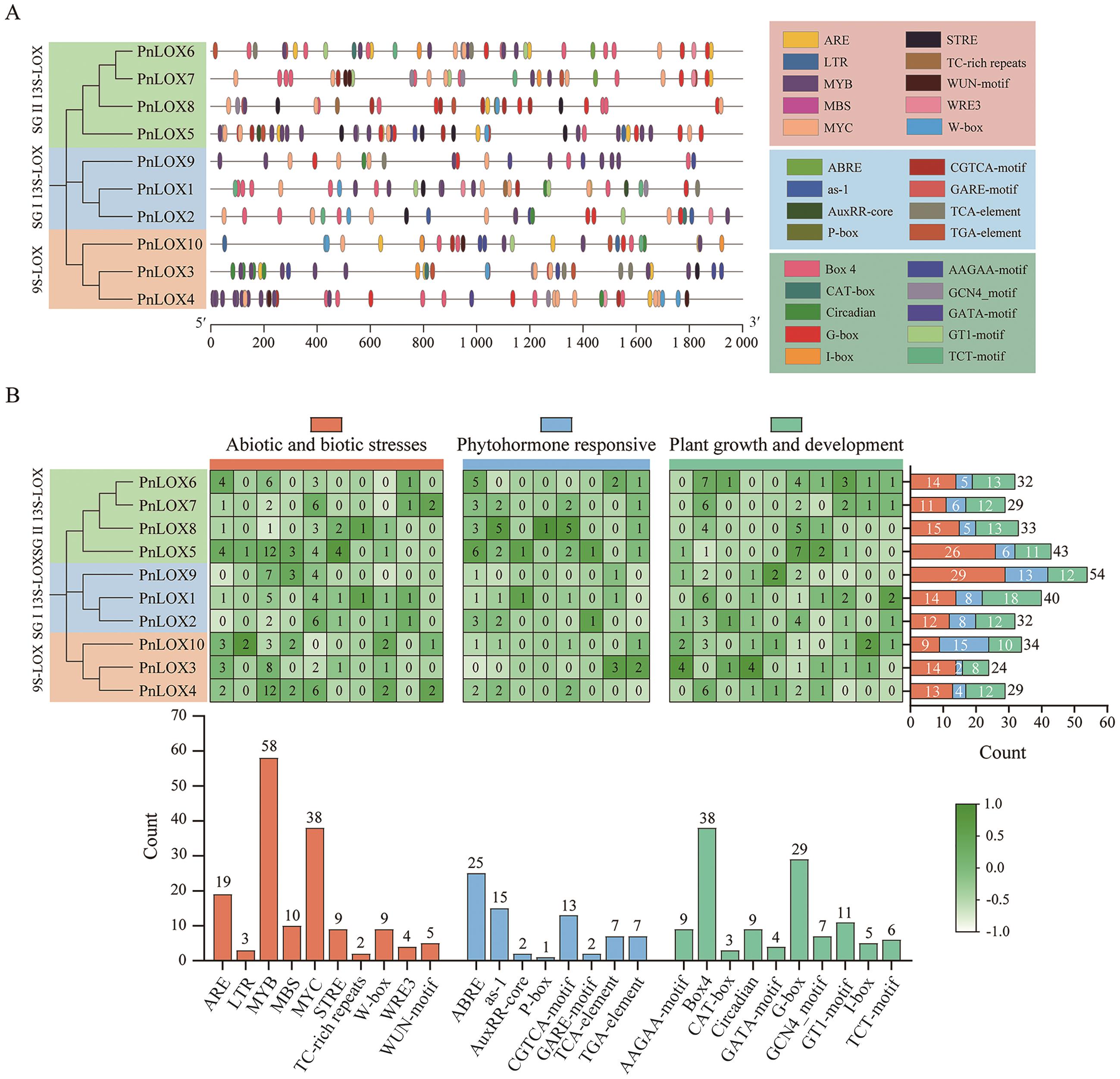
Fig. 4 Cis-acting elements in the promoter regions of PnLOX genesA: Distribution of PnLOX genes cis-acting elements on the promoter. Different colors indicate the different cis-acting elements. B: The number of cis-acting elements in PnLOX genes. The heatmap in grid and the color columns indicate the numbers of cis-acting elements
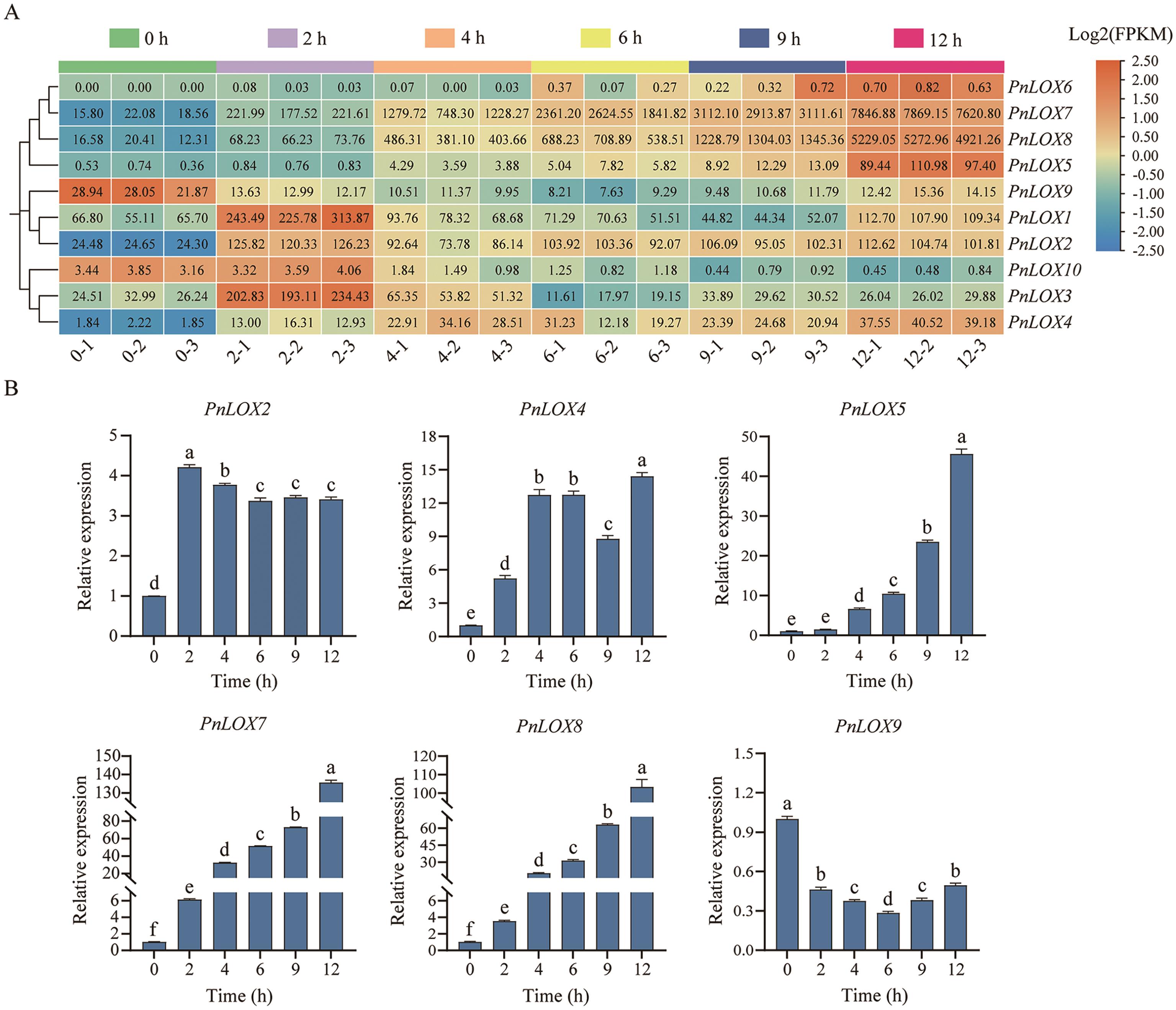
Fig. 6 Expression profiles of PnLOXs under MeJA treatmentA. Transcriptome expression heatmap of PnLOX genes after MeJA treatment. B. RT-qPCR results of 6 PnLOX genes after MeJA treatment
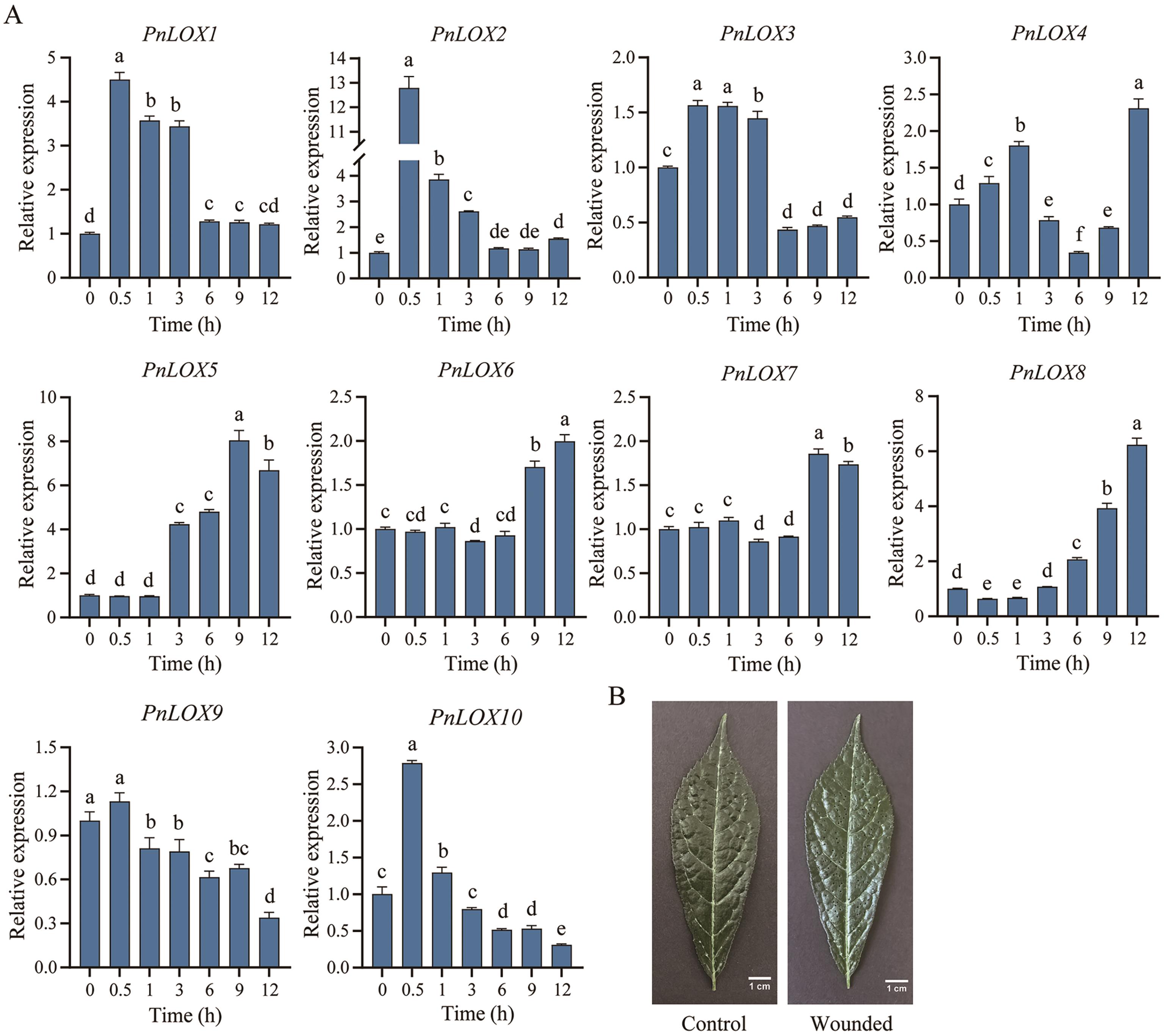
Fig. 7 Expression profiles of PnLOXs in response to wound stressA: Expressions of PnLOX genes at different times after wound stress. B: Un-wounded and wounded leaf of P. notoginseng
| [1] | Wasternack C, Feussner I. The oxylipin pathways: biochemistry and function [J]. Annu Rev Plant Biol, 2018, 69: 363-386. |
| [2] | Liavonchanka A, Feussner I. Lipoxygenases: occurrence, functions and catalysis [J]. J Plant Physiol, 2006, 163(3): 348-357. |
| [3] | Babenko LM, Shcherbatiuk MM, Skaterna TD, et al. Lipoxygenases and their metabolites in formation of plant stress tolerance [J]. UkrBiochemJ, 2017, 89(1): 5-21. |
| [4] | Fayaz M, Kundan M, Gani U, et al. Identification of Lipoxygenase gene repertoire of Cannabis sativa and functional characterization of CsLOX13 gene [J]. Plant Sci, 2023, 334: 111780. |
| [5] | Umate P. Genome-wide analysis of lipoxygenase gene family in Arabidopsis and rice [J]. Plant Signal Behav, 2011, 6(3): 335-338. |
| [6] | Sarde SJ, Kumar A, Remme RN, et al. Genome-wide identification, classification and expression of lipoxygenase gene family in pepper [J]. Plant Mol Biol, 2018, 98(4): 375-387. |
| [7] | Xu L, Zhu XX, Yi FY, et al. A genome-wide study of the lipoxygenase gene families in Medicago truncatula and Medicago sativa reveals that MtLOX24 participates in the methyl jasmonate response [J]. BMC Genom, 2024, 25(1): 195. |
| [8] | Meng Y, Liang Y, Liao BS, et al. Genome-wide identification, characterization and expression analysis of lipoxygenase gene family in Artemisia annua L [J]. Plants, 2022, 11(5): 655. |
| [9] | Upadhyay RK, Mattoo AK. Genome-wide identification of tomato (Solanum lycopersicum L.) lipoxygenases coupled with expression profiles during plant development and in response to methyl-jasmonate and wounding [J]. J Plant Physiol, 2018, 231: 318-328. |
| [10] | Zhang C, Jin YZ, Liu JY, et al. The phylogeny and expression profiles of the lipoxygenase (LOX) family genes in the melon (Cucumis melo L.) genome [J]. Sci Hortic, 2014, 170: 94-102. |
| [11] | Chauvin A, Lenglet A, Wolfender JL, et al. Paired hierarchical organization of 13-lipoxygenases in Arabidopsis [J]. Plants, 2016, 5(2): 16. |
| [12] | Chauvin A, Caldelari D, Wolfender JL, et al. Four 13-lipoxygenases contribute to rapid jasmonate synthesis in wounded Arabidopsis thaliana leaves: a role for lipoxygenase 6 in responses to long-distance wound signals [J]. New Phytol, 2013, 197(2): 566-575. |
| [13] | Zhou GX, Qi JF, Ren N, et al. Silencing OsHI-LOX makes rice more susceptible to chewing herbivores, but enhances resistance to a phloem feeder [J]. Plant J, 2009, 60(4): 638-648. |
| [14] | Sarde SJ, Bouwmeester K, Venegas-Molina J, et al. Involvement of sweet pepper CaLOX2 in jasmonate-dependent induced defence against Western flower Thrips [J]. J Integr Plant Biol, 2019, 61(10): 1085-1098. |
| [15] | Yan LH, Zhai QZ, Wei JN, et al. Role of tomato lipoxygenase D in wound-induced jasmonate biosynthesis and plant immunity to insect herbivores [J]. PLoS Genet, 2013, 9(12): e1003964. |
| [16] | Zhao C, Liu W, Yao CX, et al. AcNAC10, regulated by AcTGA07, enhances kiwifruit resistance to Pseudomonas syringae pv. actinidiae via inhibiting jasmonic acid pathway [J]. Mol Hortic, 2025, 5(1): 21. |
| [17] | Hwang IS, Hwang BK. The pepper 9-lipoxygenase gene CaLOX1 functions in defense and cell death responses to microbial pathogens [J]. Plant Physiol, 2010, 152(2): 948-967. |
| [18] | Xing QJ, Wang LX, Wu XT, et al. Red light induces powdery mildew resistance by activating the expression of CmLOX10 in oriental melon [J]. Plant Cell Environ, 2025, 48(7): 5529-5544. |
| [19] | Lv BB, Feng QQ, Shao GG, et al. The transcription factor Dof32 coordinates salvianolic acid biosynthesis and drought tolerance in Salvia miltiorrhiza [J]. Plant Physiol, 2025, 198(2): kiaf221. |
| [20] | Rahimi S, Kim YJ, Sukweenadhi J, et al. PgLOX6 encoding a lipoxygenase contributes to jasmonic acid biosynthesis and ginsenoside production in Panax ginseng [J]. J Exp Bot, 2016: erw413. |
| [21] | Shen JY, Tieman D, Jones JB, et al. A 13-lipoxygenase, TomloxC, is essential for synthesis of C5 flavour volatiles in tomato [J]. J Exp Bot, 2014, 65(2): 419-428. |
| [22] | Yuan PG, Borrego E, Park YS, et al. 9, 10-KODA, an α-ketol produced by the tonoplast-localized 9-lipoxygenase ZmLOX5, plays a signaling role in maize defense against insect herbivory [J]. Mol Plant, 2023, 16(8): 1283-1303. |
| [23] | Christensen SA, Nemchenko A, Borrego E, et al. The maize lipoxygenase, ZmLOX10, mediates green leaf volatile, jasmonate and herbivore-induced plant volatile production for defense against insect attack [J]. Plant J, 2013, 74(1): 59-73. |
| [24] | Wang T, Guo RX, Zhou GH, et al. Traditional uses, botany, phytochemistry, pharmacology and toxicology of Panax notoginseng (Burk.) F.H. Chen: a review [J]. J Ethnopharmacol, 2016, 188: 234-258. |
| [25] | Moses T, Pollier J, Almagro L, et al. Combinatorial biosynthesis of sapogenins and saponins in Saccharomyces cerevisiae using a C-16α hydroxylase from Bupleurum falcatum [J]. Proc Natl Acad Sci U S A, 2014, 111(4): 1634-1639. |
| [26] | Li JB, Bao YL, Wang ZR, et al. Research progress in diseases of Panax notoginseng [J]. Physiol Mol Plant Pathol, 2022, 121: 101878. |
| [27] | Zheng LL, Qiu BL, Su LL, et al. Panax notoginseng WRKY transcription factor 9 is a positive regulator in responding to root rot pathogen Fusarium solani [J]. Front Plant Sci, 2022, 13: 930644. |
| [28] | Tong YR, Chen K, Jiang ZQ, et al. Spatiotemporal expression analysis of jasmonic acid and saponin-related genes uncovers a potential biosynthetic regulation in Panax notoginseng [J]. J Sci Food Agric, 2024, 104(15): 9772-9781. |
| [29] | Dai HY, Zhang XK, Bi Y, et al. Improvement of Panax notoginseng saponin accumulation triggered by methyl jasmonate under arbuscular mycorrhizal fungi [J]. Front Plant Sci, 2024, 15: 1360919. |
| [30] | Yang ZJ, Liu GZ, Zhang GH, et al. The chromosome-scale high-quality genome assembly of Panax notoginseng provides insight into dencichine biosynthesis [J]. Plant Biotechnol J, 2021, 19(5): 869-871. |
| [31] | Chen CJ, Wu Y, Li JW, et al. TBtools-II: a “one for all, all for one” bioinformatics platform for biological big-data mining [J]. Mol Plant, 2023, 16(11): 1733-1742. |
| [32] | Kumar S, Stecher G, Suleski M, et al. MEGA12: molecular evolutionary genetic analysis version 12 for adaptive and green computing [J]. Mol Biol Evol, 2024, 41(12): msae263. |
| [33] | Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation [J]. Nucleic Acids Res, 2021, 49(W1): W293-W296. |
| [34] | Wang JY, Chitsaz F, Derbyshire MK, et al. The conserved domain database in 2023 [J]. Nucleic Acids Res, 2023, 51(D1): D384-D388. |
| [35] | Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers [J]. Proc Int Conf Intell Syst Mol Biol, 1994, 2: 28-36. |
| [36] | Lescot M, Déhais P, Thijs G, et al. PlantCARE, a database of plant Cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences [J]. Nucleic Acids Res, 2002, 30(1): 325-327. |
| [37] | Yang Q, Li JB, Ma WN, et al. Melatonin increases leaf disease resistance and saponin biosynthesis in Panax notogiseng [J]. J Plant Physiol, 2021, 263: 153466. |
| [38] | Ozalvo R, Cabrera J, Escobar C, et al. Two closely related members of Arabidopsis 13-lipoxygenases (13-LOXs), LOX3 and LOX4, reveal distinct functions in response to plant-parasitic nematode infection [J]. Mol Plant Pathol, 2014, 15(4): 319-332. |
| [39] | Yi R, Li YR, Shan XY. OPDA/dn-OPDA actions: biosynthesis, metabolism, and signaling [J]. Plant Cell Rep, 2024, 43(8): 206. |
| [40] | Tang HT, Lin SM, Deng JL, et al. Engineering yeast for the de novo synthesis of jasmonates [J]. Nat Synth, 2024, 3(2): 224-235. |
| [41] | Hu RB, Qi G, Kong YZ, et al. Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa [J]. BMC Plant Biol, 2010, 10(1): 145. |
| [42] | Holub EB. The arms race is ancient history in Arabidopsis, the wildflower [J]. Nat Rev Genet, 2001, 2(7): 516-527. |
| [43] | Cannon SB, Mitra A, Baumgarten A, et al. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana [J]. BMC Plant Biol, 2004, 4(1): 10. |
| [44] | Benjamin Fode TS. The Arabidopsis GRAS protein SCL14 interacts with class II TGA transcription factors and is essential for the activation of stress-inducible promoters [J]. Plant Cell, 2008, 20(11): 3122-3135. |
| [45] | Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses [J]. Annu Rev Plant Biol, 2006, 57: 781-803. |
| [46] | Christopher Johnson EB. Salicylic acid and NPR1 induce the recruitment of trans-activating TGA factors to a defense gene promoter in Arabidopsis [J]. Plant Cell, 2003, 15(8): 1846-1858. |
| [47] | Lam E, Chua NH. GT-1 binding site confers light responsive expression in transgenic tobacco [J]. Science, 1990, 248(4954): 471-474. |
| [48] | Chen Z, Chen X, Yan HW, et al. The lipoxygenase gene family in poplar: identification, classification, and expression in response to MeJA treatment [J]. PLoS One, 2015, 10(4): e0125526. |
| [49] | Verma V, Ravindran P, Kumar PP. Plant hormone-mediated regulation of stress responses [J]. BMC Plant Biol, 2016, 16(1): 86. |
| [50] | Zhang GF, Zhao F, Chen LQ, et al. Jasmonate-mediated wound signalling promotes plant regeneration [J]. Nat Plants, 2019, 5(5): 491-497. |
| [51] | Liao ZH, Wang L, Li CZ, et al. The lipoxygenase gene OsRCI-1 is involved in the biosynthesis of herbivore-induced JAs and regulates plant defense and growth in rice [J]. Plant Cell Environ, 2022, 45(9): 2827-2840. |
| [52] | Mao KM, Li CZ, Zhai HC, et al. OsRCI-1-mediated GLVs enhance rice resistance to brown planthoppers [J]. Plants, 2024, 13(11): 1494. |
| [53] | Shahzad R, Waqas M, Khan AL, et al. Foliar application of methyl jasmonate induced physio-hormonal changes in Pisum sativum under diverse temperature regimes [J]. Plant Physiol Biochem, 2015, 96: 406-416. |
| [54] | Shrestha K, Pant S, Huang YH. Genome-wide identification and classification of Lipoxygenase gene family and their roles in sorghum-aphid interaction [J]. Plant Mol Biol, 2021, 105(4): 527-541. |
| [55] | Fuminori Takahashi RY. The mitogen-activated protein kinase cascade MKK3-MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis [J]. Plant Cell, 2007, 19(3): 805-818. |
| [56] | Yang TH, Lenglet-Hilfiker A, Stolz S, et al. Jasmonate precursor biosynthetic enzymes LOX3 and LOX4 control wound-response growth restriction [J]. Plant Physiol, 2020, 184(2): 1172-1180. |
| [1] | HE Qi-chen, YANG Yang, ALIYA Waili, TANG Xin-yue, LI Zhong-xi, CHEN Yong-kun, CHEN Ling-na. Investigation into the Family Characteristics of the Lavender Copper Amine Oxidase Gene and the Role of LaCuAO1 in Bioamine Degradation [J]. Biotechnology Bulletin, 2026, 42(1): 114-124. |
| [2] | YANG Dan, JIN Ya-rong, MAO Chun-li, WANG Bi-xian, ZHANG Ya-ning, YANG Zhi-yi, ZHOU Zhi-yao, YANG Rui-ming, FAN Heng-rui, HUANG Lin-kai, YAN Hai-dong. Identification and Expression Analysis of C2H2 Gene Family in Cenchrus purpureus [J]. Biotechnology Bulletin, 2026, 42(1): 251-261. |
| [3] | GAN Chen-lu, YOU Yu-ting, XIE Han-dan, ZENG Zi-xian, ZHU Bo. Research Progress in Flavin Monooxygenases in Plants [J]. Biotechnology Bulletin, 2026, 42(1): 1-12. |
| [4] | LONG Lin-xi, ZENG Yin-ping, WANG Qian, DENG Yu-ping, GE Min-qian, CHEN Yan-zhuo, LI Xin-juan, YANG Jun, ZOU Jian. Identification of Sunflower GH3 Gene Family and Analysis of Their Function in Flower Development [J]. Biotechnology Bulletin, 2026, 42(1): 125-138. |
| [5] | LI Shan, MA Deng-hui, MA Hong-yi, YAO Wen-kong, YIN Xiao. Identification and Expression Analysis of SKP1 Gene Family in Grapevine (Vitis vinifera L.) [J]. Biotechnology Bulletin, 2025, 41(9): 147-158. |
| [6] | HUANG Guo-dong, DENG Yu-xing, CHENG Hong-wei, DAN Yan-nan, ZHOU Hui-wen, WU Lan-hua. Genome-wide Identification and Expression Analysis of the ZIP Gene Family in Soybean [J]. Biotechnology Bulletin, 2025, 41(9): 71-81. |
| [7] | CHENG Ting-ting, LIU Jun, WANG Li-li, LIAN Cong-long, WEI Wen-jun, GUO Hui, WU Yao-lin, YANG Jing-fan, LAN Jin-xu, CHEN Sui-qing. Genome-wide Identification of the Chalcone Isomerase Gene Family in Eucommia ulmoides and Analysis of Their Expression Patterns [J]. Biotechnology Bulletin, 2025, 41(9): 242-255. |
| [8] | CHENG Xue, FU Ying, CHAI Xiao-jiao, WANG Hong-yan, DENG Xin. Identification of LHC Gene Family in Setaria italica and Expression Analysis under Abiotic Stresses [J]. Biotechnology Bulletin, 2025, 41(8): 102-114. |
| [9] | HUA Wen-ping, LIU Fei, HAO Jia-xin, CHEN Chen. Identification and Expression Patterns Analysis of ADH Gene Family in Salvia miltiorrhiza [J]. Biotechnology Bulletin, 2025, 41(8): 211-219. |
| [10] | LA Gui-xiao, ZHAO Yu-long, DAI Dan-dan, YU Yong-liang, GUO Hong-xia, SHI Gui-xia, JIA Hui, YANG Tie-gang. Identification of Plasma Membrane H+-ATPase Gene Family in Safflower and Expression Analysis in Response to Low Nitrogen and Low Phosphorus Stress [J]. Biotechnology Bulletin, 2025, 41(8): 220-233. |
| [11] | HUANG Shi-yu, TIAN Shan-shan, YANG Tian-wei, GAO Man-rong, ZHANG Shang-wen. Genome-wide Identification and Expression Pattern Analysis of WRI1 Gene Family in Erythropalum scandens [J]. Biotechnology Bulletin, 2025, 41(8): 242-254. |
| [12] | REN Rui-bin, SI Er-jing, WAN Guang-you, WANG Jun-cheng, YAO Li-rong, ZHANG Hong, MA Xiao-le, LI Bao-chun, WANG Hua-jun, MENG Ya-xiong. Identification and Expression Analysis of GH17 Gene Family of Pyrenophora graminea [J]. Biotechnology Bulletin, 2025, 41(8): 146-154. |
| [13] | ZHANG Ze, YANG Xiu-li, NING Dong-xian. Identification of 4CL Gene Family in Arachis hypogaea L. and Expression Analysis in Response to Drought and Salt Stress [J]. Biotechnology Bulletin, 2025, 41(7): 117-127. |
| [14] | LI Kai-yue, DENG Xiao-xia, YIN Yuan, DU Ya-tong, XU Yuan-jing, WANG Jing-hong, YU Song, LIN Ji-xiang. Identification of LEA Gene Family and Analysis on Its Response to Aluminum Stress in Ricinus communis L. [J]. Biotechnology Bulletin, 2025, 41(7): 128-138. |
| [15] | GONG Yu-han, CHEN Lan, SHANGFANG Hui-zi, HAO Ling-ying, LIU Shuo-qian. Identification and Expression Profile Analysis of the TRB Gene Family in Tea Plant [J]. Biotechnology Bulletin, 2025, 41(7): 214-225. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||