








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (12): 190-200.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0861
Previous Articles Next Articles
ZENG Jing-jing1( ), LUO Pan-lan1, YAN Shu-jun1, ZHENG Tao2, YANG Jun-jie2, CAI Kun-xiu2, CAO Jia-yu1, ZHANG Tian-xiang2, LI Luan2, CHEN Ying1(
), LUO Pan-lan1, YAN Shu-jun1, ZHENG Tao2, YANG Jun-jie2, CAI Kun-xiu2, CAO Jia-yu1, ZHANG Tian-xiang2, LI Luan2, CHEN Ying1( )
)
Received:2025-08-09
Online:2025-12-26
Published:2026-01-06
Contact:
CHEN Ying
E-mail:15528517916@163.com;000q020057@fafu.edu.cn
ZENG Jing-jing, LUO Pan-lan, YAN Shu-jun, ZHENG Tao, YANG Jun-jie, CAI Kun-xiu, CAO Jia-yu, ZHANG Tian-xiang, LI Luan, CHEN Ying. Potential Regulatory Differences of Endogenous Hormones on Flavonoids in Anoectochilus roxburghii at Different Growth Stages Based on Multi-omics Analysis[J]. Biotechnology Bulletin, 2025, 41(12): 190-200.

Fig. 1 Anoectochilus roxburghii at different growth stagesFrom left to right, they are three-month-old, six-month-old, and ten-month-old, respectively
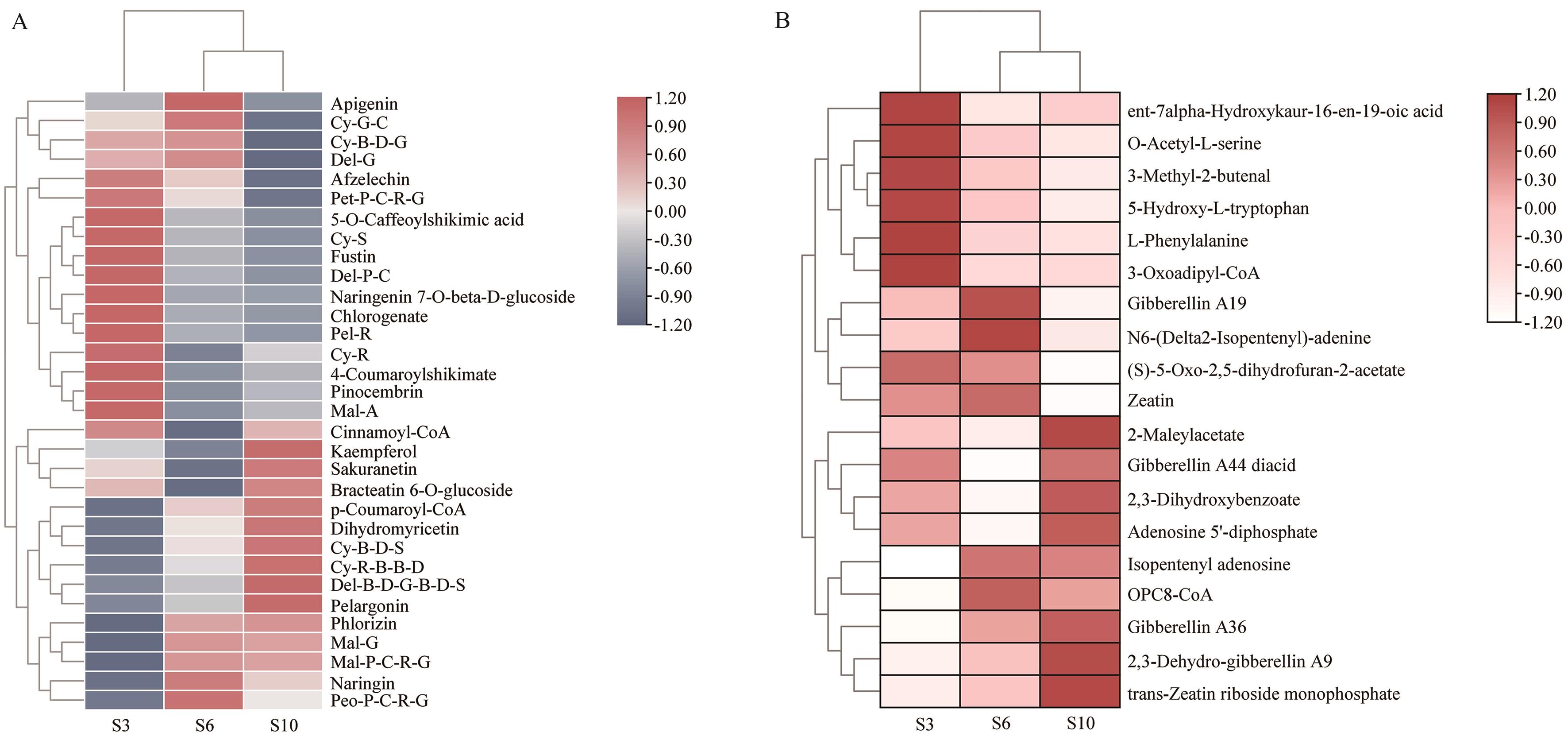
Fig. 2 Clustered heatmap of differential flavonoid and plant hormone metabolites in the leaves of A. roxburghii at various growth stagesA: Flavonoid metabolites; B: Plant hormone metabolites

Fig. 3 Hierarchical-clustering heatmap of flavonoid-synthesis-related differentially expressed genes in A. roxburghii leaves at different growth stages
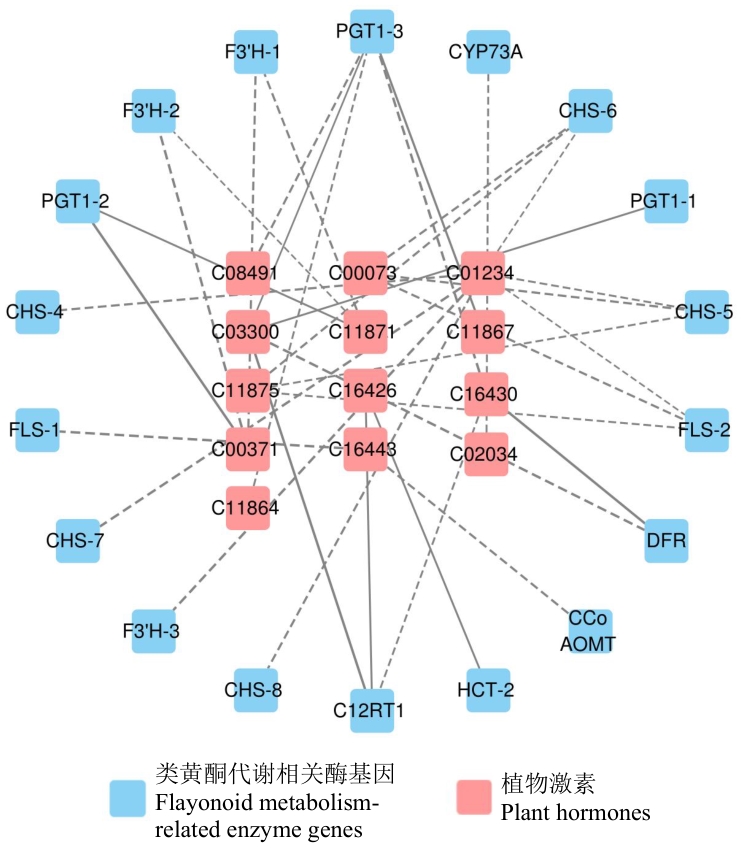
Fig. 5 Correlation network between phytohormones and flavonoid-related genes in A. roxburghii leaves at different growth stagesPlant hormone metabolites are denoted by KEGG annotation numbers. Solid lines indicate positive regulation, and dashed lines indicate negative regulation
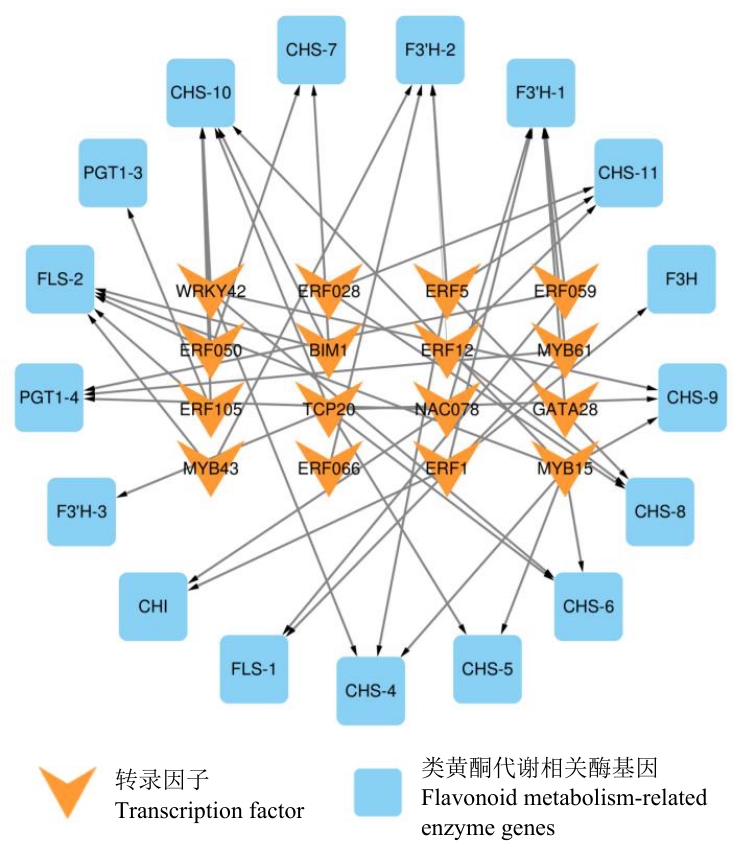
Fig. 6 Regulatory network between transcription factors and key enzyme genes of flavonoid synthesis in A. roxburghii leaves at different growth stages
| [1] | 邵清松.金线莲仿野生栽培技术[J].浙江林业,2023,(11):20-21. |
| Shao Q S. Imitation-wild cultivation techniques of Anoectochilus roxburghii [J]. Zhejiang Forestry, 2023, (11): 20-21. | |
| [2] | 林兵, 陈艺荃, 樊荣辉, 等. 花叶金线莲试管花卉培养基配方筛选 [J]. 福建农业学报, 2018, 33(12): 1275-1279. |
| Lin B, Chen YQ, Fan RH, et al. Medium formula selection for tube flowers of Anoectochilus roxburghii (wall.) lindl.Huaye [J]. Fujian J Agric Sci, 2018, 33(12): 1275-1279. | |
| [3] | 张闻婷, 梅瑜, 王继华. 珍稀药用植物金线莲研究现状与展望 [J]. 中国农学通报, 2024, 40(5): 16-26. |
| Zhang WT, Mei Y, Wang JH. Research status and prospect of precious medicinal plant Anoectochilus roxburghii [J]. Chin Agric Sci Bull, 2024, 40(5): 16-26. | |
| [4] | Ye SY, Shao QS, Zhang AL. Anoectochilus roxburghii: a review of its phytochemistry, pharmacology, and clinical applications [J]. J Ethnopharmacol, 2017, 209: 184-202. |
| [5] | 葛诗蓓, 张学宁, 韩文炎, 等. 植物类黄酮的生物合成及其抗逆作用机制研究进展 [J]. 园艺学报, 2023, 50(1): 209-224. |
| Ge SB, Zhang XN, Han WY, et al. Research progress on plant flavonoids biosynthesis and their anti-stress mechanism [J]. Acta Hortic Sin, 2023, 50(1): 209-224. | |
| [6] | Panche AN, Diwan AD, Chandra SR. Flavonoids: an overview [J]. J Nutr Sci, 2016, 5: e47. |
| [7] | Gilbert ER, Liu D. Flavonoids influence epigenetic-modifying enzyme activity: structure-function relationships and the therapeutic potential for cancer [J]. Curr Med Chem, 2010, 17(17): 1756-1768. |
| [8] | Middleton E, Kandaswami C, Theoharides TC. The effects of plant flavonoids on mammalian cells: implications for inflammation, heart disease, and cancer [J]. Pharmacol Rev, 2000, 52(4): 673-751. |
| [9] | 梁婉凤, 曾菁菁, 胡若群, 等. 转录组与代谢组分析不同生长时期金线莲类胡萝卜素的积累 [J]. 生物技术通报, 2024, 40(10): 262-274. |
| Liang WF, Zeng JJ, Hu RQ, et al. Transcriptional and metabolomic analysis of carotenoid accumulation in Anoectochilus roxburghii during different growth periods [J]. Biotechnol Bull, 2024, 40(10): 262-274. | |
| [10] | Xiao JP, Xu XY, Li MX, et al. Regulatory network characterization of anthocyanin metabolites in purple sweetpotato via joint transcriptomics and metabolomics [J]. Front Plant Sci, 2023, 14: 1030236. |
| [11] | Xiao YC, Tang YL, Huang XH, et al. Integrated transcriptomics and metabolomics analysis reveal anthocyanin biosynthesis for petal color formation in Catharanthus roseus . [J]. Agronomy, 2023, 13(9): 2290. |
| [12] | 杨运良, 苏彩虹, 李红义, 等. 综合代谢组学和转录组学分析揭示火龙果不同发育阶段的潜在颜色变化 [J]. 热带作物学报, 2022, 43(2): 225-234. |
| Yang YL, Su CH, Li HY, et al. Integrated metabolomic and transcriptomic analyses revealing potential color changes during different developmental stages of dragon fruit [J]. Chin J Trop Crops, 2022, 43(2): 225-234. | |
| [13] | Kanehisa M, Goto S. KEGG Kyoto encyclopedia of genes and genomes [J]. Nucleic Acids Res, 2000, 28(1): 27-30. |
| [14] | Ji XL, Ren J, Zhang YX, et al. Integrated analysis of the metabolome and transcriptome on anthocyanin biosynthesis in four developmental stages of Cerasus humilis peel coloration [J]. Int J Mol Sci, 2021, 22(21): 11880. |
| [15] | Chen K, Liu J, Ji RF, et al. Biogenic synthesis and spatial distribution of endogenous phytohormones and ginsenosides provide insights on their intrinsic relevance in Panax ginseng [J]. Front Plant Sci, 2019, 9: 1951. |
| [16] | 张新业, 苏彦苹, 朱姝, 等. 异源表达菊芋HtHCT基因对受体植物木质素相关生理生化指标的影响 [J]. 西北农业学报, 2022, 31(1): 63-71. |
| Zhang XY, Su YP, Zhu S, et al. Effect of heterologous expression of HtHCT gene from Helianthus tuberosus on lignin-related physiological and biochemical indicators of recipient plants [J]. Acta Agric Boreali Occidentalis Sin, 2022, 31(1): 63-71. | |
| [17] | Mao YY, Luo JJ, Cai ZP. Biosynthesis and regulatory mechanisms of plant flavonoids: a review [J]. Plants, 2025, 14(12): 1847. |
| [18] | 段玥彤, 王鹏年, 张春宝, 等. 植物黄烷酮-3-羟化酶基因研究进展 [J]. 生物技术通报, 2022, 38(6): 27-33. |
| Duan YT, Wang PN, Zhang CB, et al. Research progress in plant flavanone-3-hydroxylase gene [J]. Biotechnol Bull, 2022, 38(6): 27-33. | |
| [19] | 刘卫平, 任亚超, 杨敏生, 等. ‘冰川红叶’小檗对遮荫的生理响应及转录组分析 [J]. 植物生理学报, 2023, 59(5): 977-996. |
| Liu WP, Ren YC, Yang MS, et al. Physiological response and transcriptome analyses of Berberis thunbergii ‘Bingchuanhongye’ to shading [J]. Plant Physiol J, 2023, 59(5): 977-996. | |
| [20] | 薛晓敏. 套袋苹果果皮光照诱导后黑暗着色机理解析 [D]. 杨凌: 西北农林科技大学, 2021. |
| Xue XM. Analysis of dark coloring mechanism of bagged apple peel induced by light [D]. Yangling: Northwest A & F University, 2021. | |
| [21] | 乔军, 王利英, 刘婧, 等. 基于转录组测序的茄子萼下果色光敏相关基因表达分析 [J]. 园艺学报, 2022, 49(11): 2347-2356. |
| Qiao J, Wang LY, Liu J, et al. Expression analysis of genes related to photosensitive color under the caylx in eggplant based on transcriptome sequencing [J]. Acta Hortic Sin, 2022, 49(11): 2347-2356. | |
| [22] | 刘璐瑶, 曹倩文, 马晓鸽, 等. 不同生长期棉花叶片中类黄酮代谢物的变化与分布 [J]. 棉花学报, 2025, 37(1): 1-12. |
| Liu LY, Cao QW, Ma XG, et al. Changes and distribution of flavonoid metabolites in cotton leaves at different growth stages [J]. Cotton Sci, 2025, 37(1): 1-12. | |
| [23] | Yi DB, Zhang HN, Lai B, et al. Integrative analysis of the coloring mechanism of red Longan pericarp through metabolome and transcriptome analyses [J]. J Agric Food Chem, 2021, 69(6): 1806-1815. |
| [24] | 钱婕妤. 百日草花青素苷代谢的分子机制 [D]. 杭州: 浙江农林大学, 2022. |
| Qian JY. Molecular mechanism of anthocyanin metabolism in Zinnia elegans [D]. Hangzhou: Zhejiang A & F University, 2022. | |
| [25] | 齐昕宇. 燕子花花色成分鉴定及IlF3'H、IlF3'5'H基因克隆和表达模式研究 [D]. 哈尔滨: 东北林业大学, 2021. |
| Qi XY. Identification of perianth color components, and cloning and expressive pattern of IlF3'H, IlF3'5'H genes of iris laevigata [D]. Harbin: Northeast Forestry University, 2021. | |
| [26] | 姜丽娜. 金花茶花色形成代谢机理及关键基因功能研究 [D]. 北京: 中国林业科学研究院, 2020. |
| Jiang LN. Study on flower color formation metabolism mechanism and key genes function of Camellia nitidissima [D]. Beijing: Chinese Academy of Forestry, 2020. | |
| [27] | An Y, Li N, Zhang RL, et al. Identification and characterization of DFR gene family and cloning of candidate genes for anthocyanin biosynthesis in pepper (Capsicum annuum L.) [J]. BMC Plant Biol, 2025, 25(1): 830. |
| [28] | 李亚丽, 李欣, 肖婕, 等. 二氢黄酮醇-4-还原酶在花青素合成中的功能及调控研究进展 [J]. 西北植物学报, 2018, 38(1): 187-196. |
| Li YL, Li X, Xiao J, et al. Function and regulation characterization of dihydroflavonol 4-reductase in anthocyanin biosynthesis [J]. Acta Bot Boreali Occidentalia Sin, 2018, 38(1): 187-196. | |
| [29] | Luo P, Ning GG, Wang Z, et al. Disequilibrium of flavonol synthase and dihydroflavonol-4-reductase expression associated tightly to white vs. red color flower formation in plants [J]. Front Plant Sci, 2016, 6: 1257. |
| [30] | Zhang J, Xu HF, Wang N, et al. The ethylene response factor MdERF1B regulates anthocyanin and proanthocyanidin biosynthesis in apple [J]. Plant Mol Biol, 2018, 98(3): 205-218. |
| [31] | 伍小方, 高国应, 左倩, 等. FtMYB1转录因子调控苦荞毛状根黄酮醇合成的机理研究 [J]. 植物遗传资源学报, 2020, 21(5): 1270-1278. |
| Wu XF, Gao GY, Zuo Q, et al. Deciphering the functional basis of FtMYB1 transcription factor in flavonol biosynthesis of Tartary buckwheat hairy root [J]. J Plant Genet Resour, 2020, 21(5): 1270-1278. | |
| [32] | Erika Cavallini JTM. The phenylpropanoid pathway is controlled at different branches by a set of R2R3-MYB C2 repressors in grapevine [J]. Plant Physiol, 2015, 167(4): 1448-1470. |
| [33] | 刘中帅, 袁义杭, 张通, 等. 蓝莓转录因子VcMYB21在果实着色及幼苗UV处理中的响应 [J]. 植物生理学报, 2017, 53(1): 115-125. |
| Liu ZS, Yuan YH, Zhang T, et al. Expression characteristics of the transcription factor VcMYB21 in blueberry fruit coloration and response to UV in seedling [J]. Plant Physiol J, 2017, 53(1): 115-125. | |
| [34] | 窦立雯. 杨树转录因子MYB93负调控类黄酮和木质素合成的分子机制研究 [D]. 重庆: 西南大学, 2019. |
| Dou LW. The negative regulational molecular mechanism of the poplar transcription factor MYB93 involved in flavonoids and lignin biosynthesis [D]. Chongqing: Southwest University, 2019. | |
| [35] | 胡若群, 曾菁菁, 梁婉凤, 等. 转录组和代谢组联合分析探究不同遮光条件下金线莲类胡萝卜素合成代谢机制 [J]. 生物技术通报, 2025, 41(5): 231-243. |
| Hu RQ, Zeng JJ, Liang WF, et al. Integrated transcriptome and metabolome analysis to explore the carotenoid synthesis and metabolism mechanism in Anoectochilus roxburghii under different shading conditions [J]. Biotechnol Bull, 2025, 41(5): 231-243. | |
| [36] | 何磊, 潘秋红. 激素调控葡萄果实类黄酮代谢的研究进展 [J]. 热带生物学报, 2016, 7(4): 522-529. |
| He L, Pan QH. Research advances in regulation of flavonoid metabolism by plant hormones in grape berry [J]. J Trop Biol, 2016, 7(4): 522-529. | |
| [37] | 周琳, 袁梦, 齐宇, 等. 牡丹花青素苷合成关键基因PsDFR启动子活性分析 [J]. 林业科学研究, 2024, 37(2): 16-26. |
| Zhou L, Yuan M, Qi Y, et al. Promoter functional analysis of the key gene PsDFR involved in Paeonia suffruticosa anthocyanin biosynthesis [J]. For Res, 2024, 37(2): 16-26. |
| [1] | LIU Jian-guo, LIU Ge-er, GUO Ying-xin, WANG Bin, WANG Yu-kun, LU Jin-feng, HUANG Wen-ting, ZHU Yun-na. Integrate Transcriptomic and Metabolomic Analysis of Fruits Quality Differences between ‘Guiyou No. 1’ and ‘Shatianyou’ Pomelo (Citrus maxima) [J]. Biotechnology Bulletin, 2025, 41(9): 168-181. |
| [2] | LIU Ze-zhou, DUAN Nai-bin, YUE Li-xin, WANG Qing-hua, YAO Xing-hao, GAO Li-min, KONG Su-ping. Analysis of Wax Components and Screening of Wax-deficient Gene Ggl-1 in Garlic (Allium sativum L.) [J]. Biotechnology Bulletin, 2025, 41(9): 219-231. |
| [3] | YAN Meng-yang, LIANG Xiao-yang, DAI Jun-ang, ZHANG Yan, GUAN Tuan, ZHANG Hui, LIU Liang-bo, SUN Zhi-hua. Screening of Amoxicillin-degrading Bacteria and Study on Its Degradation Mechanisms [J]. Biotechnology Bulletin, 2025, 41(9): 314-325. |
| [4] | BAI Yu-guo, LI Wan-di, LIANG Jian-ping, SHI Zhi-yong, LU Geng-long, LIU Hong-jun, NIU Jing-ping. Growth-promoting Mechanism of Trichoderma harzianum T9131 on Astragalus membranaceus Seedlings [J]. Biotechnology Bulletin, 2025, 41(8): 175-185. |
| [5] | ZHANG Yue, BI Yu, MU Xue-nan, ZHENG Zi-wei, WANG Zhi-gang, XU Wei-hui. Biocontrol Characteristics of Strain JB7 against Fusarium graminearum [J]. Biotechnology Bulletin, 2025, 41(7): 261-271. |
| [6] | LI Cheng-hua, DOU Fei-fei, REN Yu-zhao, LIU Cai-xia, LIU Feng-lou, WANG Zhang-jun, LI Qing-feng. Effect of Exogenous Salicylic Acid on Wheat Infested with Blumeria graminis f. sp. tritici and Its Transcriptome Analysis [J]. Biotechnology Bulletin, 2025, 41(7): 272-280. |
| [7] | GUO Xiu-juan, FENG Yu, WU Rui-xiang, WANG Li-qin, YANG Jian-chun. Transcriptome Analysis of the Effect of Ca 2+ Treatment on the Seed Germination of Flax [J]. Biotechnology Bulletin, 2025, 41(7): 139-149. |
| [8] | WANG Yue-chen, HAN Xin-qi, WEI Wen-min, CUI Zhao-lan, LUO Yang-mei, CHEN Peng-ru, WANG Hai-gang, LIU long-long, ZHANG Li, WANG Lun. Biological Basis Study for Grain Shattering in Proso Millet and Identification of Genes Regulating Grain Shattering [J]. Biotechnology Bulletin, 2025, 41(7): 164-171. |
| [9] | HU Ruo-qun, ZENG Jing-jing, LIANG Wan-feng, CAO Jia-yu, HUANG Xiao-wei, LIANG Xiao-ying, QIU Ming-yue, CHEN Ying. Integrated Transcriptome and Metabolome Analysis to Explore the Carotenoid Synthesis and Metabolism Mechanism in Anoectochilus roxburghii under Different Shading Conditions [J]. Biotechnology Bulletin, 2025, 41(5): 231-243. |
| [10] | YANG Chao-jie, ZHANG Lan, CHEN Hong, HUANG Juan, SHI Tao-xiong, ZHU Li-wei, CHEN Qing-fu, LI Hong-you, DENG Jiao. Functional Identification of the Transcription Factor Gene FtbHLH3 in Regulating Flavonoid Biosynthesis in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2025, 41(4): 134-144. |
| [11] | HUANG Jin-heng, HUANG Xi, ZHANG Jia-yan, ZHOU Xin-yu, LIAO Pei-ran, YANG Quan. Identification and Expression Analysis of the C3H Gene Family in Grona styracifolia across Different Varieties [J]. Biotechnology Bulletin, 2025, 41(4): 243-255. |
| [12] | ZHAO Chang-yan, LIU Yan-tao, JIA Xiu-ping, LIU Sheng-li, LEI Zhong-hua, WANG Peng, ZHU Zhi-feng, DONG Hong-ye, LYU Zeng-shuai, DUAN Wei, WAN Su-mei. Research Progress in the Effect of Melatonin on Crop Physiological Mechanism under Saline-alkali Stress [J]. Biotechnology Bulletin, 2025, 41(2): 18-29. |
| [13] | LI Yan-wei, YANG Yan-yan, SUN Ya-ling, HUO Yu-meng, WANG Zhen-bao, LIU Bing-jiang. Regulation Mechanism of Plant Hormones Related to Onion Bulb Enlargement and Development Based on Transcriptome Analysis [J]. Biotechnology Bulletin, 2025, 41(2): 187-201. |
| [14] | YANG Yong, CAO Rui, KANG Xiao-xiao, LIU Jing, WANG Xuan, ZHANG Hai-e. Identification and Expression Analysis of 13 Gene Families in the Chestnut Flavonoid Synthesis Pathway [J]. Biotechnology Bulletin, 2025, 41(2): 270-283. |
| [15] | YANG Tao, LI Lin, MO Xiao-lian, CHEN Xiao-long, WANG Jian, HUANG Yuan, ZHAO Jie-hong, ZOU Jie. Functional Study of DoDELLA2 in Dendrobium officinale Kimura et Migo [J]. Biotechnology Bulletin, 2025, 41(12): 240-253. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||