








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (6): 1-12.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0450
Received:2021-04-07
Online:2021-06-26
Published:2021-07-08
Contact:
ZHOU Qian
E-mail:tangdie_solab@163.com;zhouq02@pcl.ac.cn
TANG Die, ZHOU Qian. Research Advances in Plant Genome Assembly[J]. Biotechnology Bulletin, 2021, 37(6): 1-12.
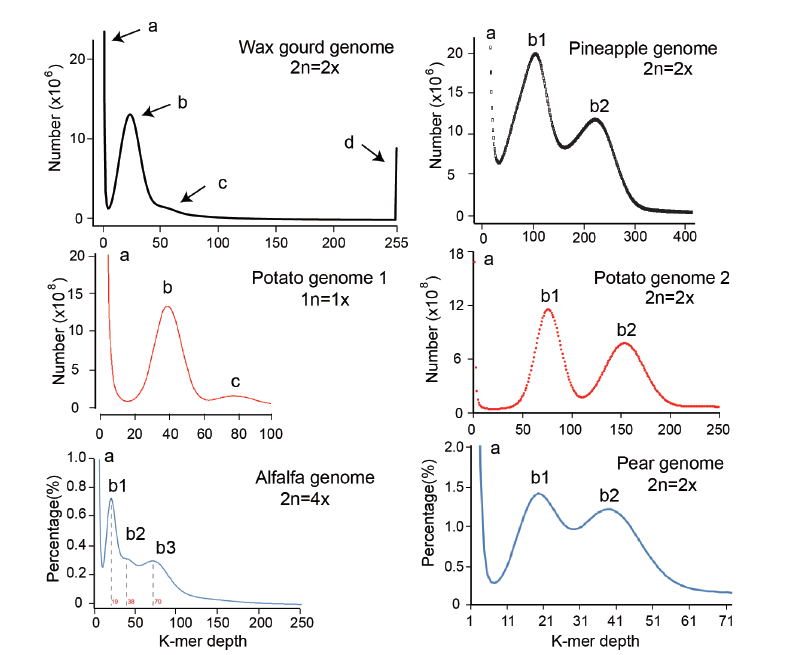
Fig. 1 K-mer volume histograms of illumina sequencing data from several plant genomes a:The first peak,at the depth at 1-2,is derived from sequencing error. b:The second peak,is the main peak in haploid genome or homozygous diploid genome. c-d:The peak c,is composed of the repetitive k-mers with relative low copy number while the right-most peak d,is composed of the highly repetitive k-mers. In heterozygous diploid genome,the peak b1 contains k-mers derived from heterozygous regions and the peak b2 contains k-mers derived from homozygous regions. The depth of peak b1 is only half of the average sequencing depth. In heterozygous autotetraploid genome,both of the peak b1 and b2 present heterozygous k-mers and the peak b3 presents homozygous k-mers. This figure is modified based on the references[4,5,6,7,8,9]
| 植物物种 Plant | 基因组大小 Genome size | 组装数据a Sequencing data | 组装策略 Assembly strategy | 组装软件 Assembly software | 纠错软件 Polish software | 组装大小 Assembly size | Contig N50 | 挂载染色体 Construct chromosomes |
|---|---|---|---|---|---|---|---|---|
| 马铃薯 DMv2.1[ | 844 Mb | Illumina,454, Sanger,共114X | 二代组装 | SOAPdenovo[ | 无 | 773 Mb | 32 kb | 物理图谱 遗传图谱 |
| 番茄 SL2.40[ | 900 Mb | 454 30X Sanger 5.2X SOLiD 140X Illumina 70X | 二代组装 | Newbler[ | 无 | 760 Mb | 87 kb | 物理图谱 遗传图谱 |
| 海草[ | 200 Mb | Illumina 50X | 二代组装 | Arachne[ | 无 | 204 Mb | 80 kb | 无 |
| 辣椒[ | 3.5 Gb | Illumina 56X | 二代组装 | Supernova[ | 无 | 3.2 Gb | 123 kb | 无 |
| 冬瓜[ | 1.03 Gb | Illumina 40X PacBio 15X | 二代组装 三代补洞 | ALLPathsLG[ | 无 | 913 Mb | 68 kb | 遗传图谱 |
| 苹果[ | 651 Mb | Illumina 80X Pacbio 35X | 二代三代 混合组装 | SOAPdenovo DBG2OLC[ | Pilon[ | 625 Mb | 620 kb | 光学图谱 |
| 野生番茄[ | 1-1.2 Gb | Illumina 35X Nanopore 100X | 三代组装 | Canu[ SMARTdenovo[ | Racon[ | 915 Mb | 2.5 Mb | 无 |
| 向日葵[ | 3.6 Gb | PacBio 100X | 三代组装 | PBcR[ | Quiver[ | 3.1 Gb | 495 kb | 遗传图谱 |
| 月季[ | 560 Mb | Illumina 150X PacBio 80X | 三代组装 | til-r,Falcon[ Canu | Quiver Pilon | 515 Mb | 24 Mb | 遗传图谱 Hi-C |
| 番茄 SL4.0[ | 900 Mb | Illumina 100X PacBio 80X | 三代组装 | Canu | Arrow Pilon | 785 Mb | 5.5 Mb | Hi-C |
| 马铃薯DMv6.1[ | 844 Mb | Illumina 80X Nanopore 45X | 三代组装 | Flye[ | Racon Nanopolish Pilon | 742 Mb | 17.3 Mb | Hi-C |
Table1 Assembly strategies and results of several plant genomes
| 植物物种 Plant | 基因组大小 Genome size | 组装数据a Sequencing data | 组装策略 Assembly strategy | 组装软件 Assembly software | 纠错软件 Polish software | 组装大小 Assembly size | Contig N50 | 挂载染色体 Construct chromosomes |
|---|---|---|---|---|---|---|---|---|
| 马铃薯 DMv2.1[ | 844 Mb | Illumina,454, Sanger,共114X | 二代组装 | SOAPdenovo[ | 无 | 773 Mb | 32 kb | 物理图谱 遗传图谱 |
| 番茄 SL2.40[ | 900 Mb | 454 30X Sanger 5.2X SOLiD 140X Illumina 70X | 二代组装 | Newbler[ | 无 | 760 Mb | 87 kb | 物理图谱 遗传图谱 |
| 海草[ | 200 Mb | Illumina 50X | 二代组装 | Arachne[ | 无 | 204 Mb | 80 kb | 无 |
| 辣椒[ | 3.5 Gb | Illumina 56X | 二代组装 | Supernova[ | 无 | 3.2 Gb | 123 kb | 无 |
| 冬瓜[ | 1.03 Gb | Illumina 40X PacBio 15X | 二代组装 三代补洞 | ALLPathsLG[ | 无 | 913 Mb | 68 kb | 遗传图谱 |
| 苹果[ | 651 Mb | Illumina 80X Pacbio 35X | 二代三代 混合组装 | SOAPdenovo DBG2OLC[ | Pilon[ | 625 Mb | 620 kb | 光学图谱 |
| 野生番茄[ | 1-1.2 Gb | Illumina 35X Nanopore 100X | 三代组装 | Canu[ SMARTdenovo[ | Racon[ | 915 Mb | 2.5 Mb | 无 |
| 向日葵[ | 3.6 Gb | PacBio 100X | 三代组装 | PBcR[ | Quiver[ | 3.1 Gb | 495 kb | 遗传图谱 |
| 月季[ | 560 Mb | Illumina 150X PacBio 80X | 三代组装 | til-r,Falcon[ Canu | Quiver Pilon | 515 Mb | 24 Mb | 遗传图谱 Hi-C |
| 番茄 SL4.0[ | 900 Mb | Illumina 100X PacBio 80X | 三代组装 | Canu | Arrow Pilon | 785 Mb | 5.5 Mb | Hi-C |
| 马铃薯DMv6.1[ | 844 Mb | Illumina 80X Nanopore 45X | 三代组装 | Flye[ | Racon Nanopolish Pilon | 742 Mb | 17.3 Mb | Hi-C |
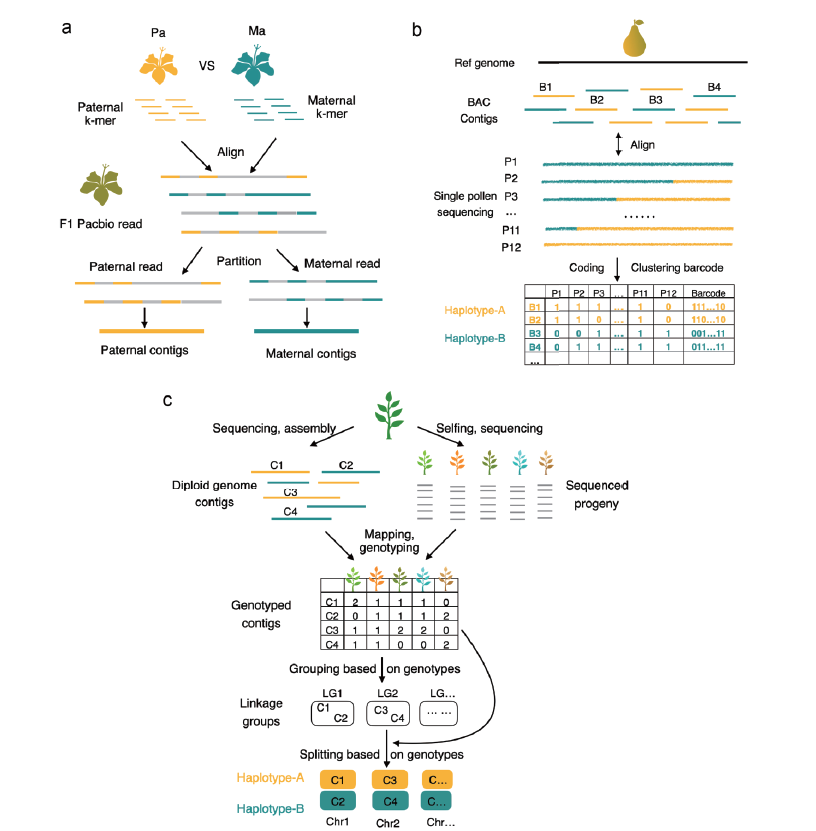
Fig. 2 Three assembly and genotyping strategies of genome in plants a:The trio-sequencing based genotyping strategy Triobin[41]. The sequencing reads of F1 hybrid was firstly partitioned into paternal and maternal sets using the parental unique K-mers,then assembled separately into two haplotypes. b:Genotyping based on haploid population[44]. To phase the pre-assembled BAC clones,12 pollen cells were sequenced individually and the alignment results between each BAC and each pollen cell were encoded into a 12-bit binary barcode. In this approach,the BAC clones could be replaced with the high-accuracy and long segment such as HiFi reads or assembled contigs,etc. c:Genotyping based on selfing segregation population[7]. Firstly,the diploid contigs were de novo assembled,and a segregation population was sequenced to identify the genotypes of contigs. Then,the genetic map was constructed to identify different chromosomes. Lastly,the contigs belonging to same chromosome were partitioned into different haplotypes based on their genotypes’ similarity

Fig. 3 Three approaches of assembling pan-genome a: Iterative assembly pan-genome. Construction of pan-genome by mapping reads to a reference genome,the unaligned reads were then assembled into novel contigs and they were iteratively added to the reference genome. b: De novo assembly pan-genome. Multiple genomes were assembled and annotated,and pan-gene clusters were predicted using clustering algorithm. Gene-clusters were further cataloged to core- and dispensable-sets according to the cluster frequency among all samples. The pan- and core-genome curves were fitted using nonlinear models. c: Graph-based genome. A pan-genome graph can be constructed by integrating variations to a reference genome. Grey box indicated the alternative path differing from reference genome. Right side showed the actual structure of 2 regions in graph-based genome
| [1] |
Simpson JT, Pop M. The theory and practice of genome sequence assembly[J]. Annu Rev Genom Hum Genet, 2015, 16(1):153-172.
doi: 10.1146/annurev-genom-090314-050032 URL |
| [2] | 高胜寒, 禹海英, 吴双阳, 等. 复杂基因组测序技术研究进展[J]. 遗传, 2018, 40(11):944-963. |
| Gao SH, Yu HY, Wu SY, et al. Advances of sequencing and assembling technologies for complex genomes[J]. Hereditas, 2018, 40(11):944-963. | |
| [3] | 杨焕明. 基因组学[M]. 北京: 科学出版社, 2016. |
| Yang HM. Genomics[M]. Beijing: Science Press, 2016. | |
| [4] |
Xie D, Xu Y, Wang J, et al. The wax gourd genomes offer insights into the genetic diversity and ancestral cucurbit karyotype[J]. Nat Commun, 2019, 10(1):5158.
doi: 10.1038/s41467-019-13185-3 URL |
| [5] |
Ming R, VanBuren R, Wai CM, et al. The pineapple genome and the evolution of CAM photosynjournal[J]. Nat Genet, 2015, 47(12):1435-1442.
doi: 10.1038/ng.3435 pmid: 26523774 |
| [6] |
Potato Genome Sequencing Consortium Xu X, Pan S, et al. Genome sequence and analysis of the Tuber crop potato[J]. Nature, 2011, 475(7355):189-195.
doi: 10.1038/nature10158 URL |
| [7] |
Zhou Q, Tang D, Huang W, et al. Haplotype-resolved genome analyses of a heterozygous diploid potato[J]. Nat Genet, 2020, 52(10):1018-1023.
doi: 10.1038/s41588-020-0699-x URL |
| [8] |
Chen H, Zeng Y, Yang Y, et al. Allele-aware chromosome-level genome assembly and efficient transgene-free genome editing for the autotetraploid cultivated alfalfa[J]. Nat Commun, 2020, 11(1):2494.
doi: 10.1038/s41467-020-16338-x URL |
| [9] |
Wu J, Wang Z, Shi Z, et al. The genome of the pear(Pyrus bretschneideri Rehd. )[J]. Genome Res, 2013, 23(2):396-408.
doi: 10.1101/gr.144311.112 URL |
| [10] |
Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution[J]. Nature, 2012, 485(7400):635-641.
doi: 10.1038/nature11119 URL |
| [11] |
Pham GM, Hamilton JP, Wood JC, et al. Construction of a chromosome-scale long-read reference genome assembly for potato[J]. Gigascience, 2020, 9(9):giaa100.
doi: 10.1093/gigascience/giaa100 URL |
| [12] | Hosmani PS, Flores-Gonzalez M, van de Geest H, et al. An improved de novo assembly and annotation of the tomato reference genome using single-molecule sequencing, Hi-C proximity ligation and optical maps[J]. bioRxiv, 2019. DOI: 10.1101/767764. |
| [13] |
Burton JN, Adey A, Patwardhan RP, et al. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions[J]. Nat Biotechnol, 2013, 31(12):1119-1125.
doi: 10.1038/nbt.2727 URL |
| [14] |
Jiao WB, Accinelli GG, Hartwig B, et al. Improving and correcting the contiguity of long-read genome assemblies of three plant species using optical mapping and chromosome conformation capture data[J]. Genome Res, 2017, 27(5):778-786.
doi: 10.1101/gr.213652.116 URL |
| [15] |
Belser C, Istace B, Denis E, et al. Chromosome-scale assemblies of plant genomes using nanopore long reads and optical maps[J]. Nat Plants, 2018, 4(11):879-887.
doi: 10.1038/s41477-018-0289-4 URL |
| [16] |
Luo R, Liu B, Xie Y, et al. SOAPdenovo2:an empirically improved memory-efficient short-read de novo assembler[J]. Gigascience, 2012, 1(1):18.
doi: 10.1186/2047-217X-1-18 URL |
| [17] |
Margulies M, Egholm M, Altman WE, et al. Genome sequencing in microfabricated high-density picolitre reactors[J]. Nature, 2005, 437(7057):376-380.
pmid: 16056220 |
| [18] |
Olsen JL, Rouzé P, Verhelst B, et al. The genome of the seagrass Zostera marina reveals angiosperm adaptation to the sea[J]. Nature, 2016, 530(7590):331-335.
doi: 10.1038/nature16548 URL |
| [19] |
Jaffe DB, Butler J, Gnerre S, et al. Whole-genome sequence assembly for mammalian genomes:Arachne 2[J]. Genome Res, 2003, 13(1):91-96.
doi: 10.1101/gr.828403 URL |
| [20] |
Hulse-Kemp AM, Maheshwari S, Stoffel K, et al. Reference quality assembly of the 3. 5-Gb genome of Capsicum annuum from a single linked-read library[J]. Hortic Res, 2018, 5:4.
doi: 10.1038/s41438-017-0011-0 URL |
| [21] |
Weisenfeld NI, Kumar V, Shah P, et al. Direct determination of diploid genome sequences[J]. Genome Res, 2017, 27(5):757-767.
doi: 10.1101/gr.214874.116 pmid: 28381613 |
| [22] |
Butler J, MacCallum I, Kleber M, et al. ALLPATHS:de novo assembly of whole-genome shotgun microreads[J]. Genome Res, 2008, 18(5):810-820.
doi: 10.1101/gr.7337908 URL |
| [23] |
Daccord N, Celton JM, Linsmith G, et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development[J]. Nat Genet, 2017, 49(7):1099-1106.
doi: 10.1038/ng.3886 URL |
| [24] |
Ye C, Hill CM, Wu S, et al. DBG2OLC:efficient assembly of large genomes using long erroneous reads of the third generation sequencing technologies[J]. Sci Rep, 2016, 6:31900.
doi: 10.1038/srep31900 URL |
| [25] |
Walker BJ, Abeel T, Shea T, et al. Pilon:an integrated tool for comprehensive microbial variant detection and genome assembly improvement[J]. PLoS One, 2014, 9(11):e112963.
doi: 10.1371/journal.pone.0112963 URL |
| [26] |
Schmidt MHW, Vogel A, Denton AK, et al. De novo assembly of a new Solanum pennellii accession using nanopore sequencing[J]. Plant Cell, 2017, 29(10):2336-2348.
doi: 10.1105/tpc.17.00521 URL |
| [27] |
Koren S, Walenz BP, Berlin K, et al. Canu:scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation[J]. Genome Res, 2017, 27(5):722-736.
doi: 10.1101/gr.215087.116 URL |
| [28] | Liu HL, Wu SG, Li AL, Ruan J. SMARTdenovo:A de novo assembler using long noisy reads[J/OL]. Preprints, 2020,2020090207. |
| [29] |
Vaser R, Sović I, Nagarajan N, et al. Fast and accurate de novo genome assembly from long uncorrected reads[J]. Genome Res, 2017, 27(5):737-746.
doi: 10.1101/gr.214270.116 URL |
| [30] |
Loman NJ, Quick J, Simpson JT. A complete bacterial genome assembled de novo using only nanopore sequencing data[J]. Nat Methods, 2015, 12(8):733-735.
doi: 10.1038/nmeth.3444 URL |
| [31] |
Badouin H, Gouzy J, Grassa CJ, et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution[J]. Nature, 2017, 546(7656):148-152.
doi: 10.1038/nature22380 pmid: 28538728 |
| [32] |
Berlin K, Koren S, Chin CS, et al. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing[J]. Nat Biotechnol, 2015, 33(6):623-630.
doi: 10.1038/nbt.3238 URL |
| [33] |
Chin CS, Alexander DH, Marks P, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data[J]. Nat Methods, 2013, 10(6):563-569.
doi: 10.1038/nmeth.2474 URL |
| [34] |
Raymond O, Gouzy J, Just J, et al. The Rosa genome provides new insights into the domestication of modern roses[J]. Nat Genet, 2018, 50(6):772-777.
doi: 10.1038/s41588-018-0110-3 URL |
| [35] |
Chin CS, Peluso P, Sedlazeck FJ, et al. Phased diploid genome assembly with single-molecule real-time sequencing[J]. Nat Methods, 2016, 13(12):1050-1054.
doi: 10.1038/NMETH.4035 |
| [36] |
Kolmogorov M, Yuan J, Lin Y, et al. Assembly of long, error-prone reads using repeat graphs[J]. Nat Biotechnol, 2019, 37(5):540-546.
doi: 10.1038/s41587-019-0072-8 pmid: 30936562 |
| [37] |
Kaper F, Swamy S, Klotzle B, et al. Whole-genome haplotyping by dilution, amplification, and sequencing[J]. PNAS, 2013, 110(14):5552-5557.
doi: 10.1073/pnas.1218696110 URL |
| [38] |
Amini S, Pushkarev D, Christiansen L, et al. Haplotype-resolved whole-genome sequencing by contiguity-preserving transposition and combinatorial indexing[J]. Nat Genet, 2014, 46(12):1343-1349.
doi: 10.1038/ng.3119 URL |
| [39] |
Mostovoy Y, Levy-Sakin M, Lam J, et al. A hybrid approach for de novo human genome sequence assembly and phasing[J]. Nat Methods, 2016, 13(7):587-590.
doi: 10.1038/nmeth.3865 pmid: 27159086 |
| [40] |
Fan YN, Sahu SK, Yang T, et al. Dissecting the genome of star fruit(Averrhoa carambola L.)[J]. Hortic Res, 2020, 7(1):1-10.
doi: 10.1038/s41438-019-0222-7 URL |
| [41] |
Koren S, Rhie A, Walenz BP, et al. De novo assembly of haplotype-resolved genomes with trio binning[J]. Nat Biotechnol, 2018, 36(12):1174-1182.
doi: 10.1038/nbt.4277 URL |
| [42] |
Cheng H, Concepcion GT, Feng X, et al. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm[J]. Nat Methods, 2021, 18(2):170-175.
doi: 10.1038/s41592-020-01056-5 URL |
| [43] |
Garg S, Fungtammasan A, Carroll A, et al. Chromosome-scale, haplotype-resolved assembly of human genomes[J]. Nat Biotechnol, 2021, 39(3):309-312.
doi: 10.1038/s41587-020-0711-0 URL |
| [44] |
Shi D, Wu J, Tang H, et al. Single-pollen-cell sequencing for gamete-based phased diploid genome assembly in plants[J]. Genome Res, 2019, 29(11):1889-1899.
doi: 10.1101/gr.251033.119 URL |
| [45] |
Schnable PS, Ware D, Fulton RS, et al. The B73 maize genome:complexity, diversity, and dynamics[J]. Science, 2009, 326(5956):1112-1115.
doi: 10.1126/science.1178534 URL |
| [46] |
Jiao Y, Peluso P, Shi J, et al. Improved maize reference genome with single-molecule technologies[J]. Nature, 2017, 546(7659):524-527.
doi: 10.1038/nature22971 URL |
| [47] |
Neale DB, Wegrzyn JL, Stevens KA, et al. Decoding the massive genome of loblolly pine using haploid DNA and novel assembly strategies[J]. Genome Biol, 2014, 15(3):R59.
doi: 10.1186/gb-2014-15-3-r59 URL |
| [48] |
Nystedt B, Street NR, Wetterbom A, et al. The Norway spruce genome sequence and conifer genome evolution[J]. Nature, 2013, 497(7451):579-584.
doi: 10.1038/nature12211 pmid: 23698360 |
| [49] |
Guan R, Zhao Y, Zhang H, et al. Draft genome of the living fossil Ginkgo biloba[J]. Gigascience, 2016, 5(1):49.
pmid: 27871309 |
| [50] |
Gan X, Stegle O, Behr J, et al. Multiple reference genomes and transcriptomes for Arabidopsis thaliana[J]. Nature, 2011, 477(7365):419-423.
doi: 10.1038/nature10414 URL |
| [51] |
Yu J, Hu S, Wang J, et al. A draft sequence of the rice genome(Oryza sativa L. ssp. indica)[J]. Science, 2002, 296(5565):79-92.
doi: 10.1126/science.1068037 URL |
| [52] | Liu HL, Wang XB, Wang GB, et al. The nearly complete genome of Ginkgo biloba illuminates gymnosperm evolution[J]. Nature Plants, 2021. (accepted) |
| [53] |
Sun X, Zhu S, Li N, et al. A chromosome-level genome assembly of garlic(Allium sativum)provides insights into genome evolution and allicin biosynjournal[J]. Mol Plant, 2020, 13(9):1328-1339.
doi: 10.1016/j.molp.2020.07.019 URL |
| [54] | The International Wheat Genome Sequencing Consortium. Shifting the limits in wheat research and breeding using a fully annotated reference genome[J]. Science, 2018, 361:1-13. |
| [55] |
Chalhoub B, Denoeud F, Liu SY, et al. Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome[J]. Science, 2014, 345(6199):950-953.
doi: 10.1126/science.1253435 URL |
| [56] |
Bertioli DJ, Cannon SB, Froenicke L, et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut[J]. Nat Genet, 2016, 48(4):438-446.
doi: 10.1038/ng.3517 pmid: 26901068 |
| [57] | Yin DM, Ji CM, Ma XL, et al. Genome of an allotetraploid wild peanut Arachis monticola:a de novo assembly[J]. Gigascience, 2018, 7(6):giy066. |
| [58] |
Li F, Fan G, Lu C, et al. Genome sequence of cultivated Upland cotton(Gossypium hirsutum TM-1)provides insights into genome evolution[J]. Nat Biotechnol, 2015, 33(5):524-530.
doi: 10.1038/nbt.3208 URL |
| [59] |
Wang M, Tu L, Yuan D, et al. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense[J]. Nat Genet, 2019, 51(2):224-229.
doi: 10.1038/s41588-018-0282-x URL |
| [60] |
Huang G, Wu Z, Percy RG, et al. Genome sequence of Gossypium herbaceum and genome updates of Gossypium arboreum and Gossypium hirsutum provide insights into cotton A-genome evolution[J]. Nat Genet, 2020, 52(5):516-524.
doi: 10.1038/s41588-020-0607-4 pmid: 32284579 |
| [61] |
Yang J, Moeinzadeh MH, Kuhl H, et al. Haplotype-resolved sweet potato genome traces back its hexaploidization history[J]. Nat Plants, 2017, 3(9):696-703.
doi: 10.1038/s41477-017-0002-z pmid: 28827752 |
| [62] |
Zhang J, Zhang X, Tang H, et al. Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L[J]. Nat Genet, 2018, 50(11):1565-1573.
doi: 10.1038/s41588-018-0237-2 URL |
| [63] |
Zhang X, Zhang S, Zhao Q, et al. Assembly of allele-aware, chromosomal-scale autopolyploid genomes based on Hi-C data[J]. Nat Plants, 2019, 5(8):833-845.
doi: 10.1038/s41477-019-0487-8 URL |
| [64] |
Zhang XT, Wu RX, Wang YB, et al. Unzipping haplotypes in diploid and polyploid genomes[J]. Comput Struct Biotechnol J, 2020, 18:66-72.
doi: 10.1016/j.csbj.2019.11.011 URL |
| [65] |
Wang W, Mauleon R, Hu Z, et al. Genomic variation in 3, 010 diverse accessions of Asian cultivated rice[J]. Nature, 2018, 557(7703):43-49.
doi: 10.1038/s41586-018-0063-9 URL |
| [66] |
Zhao Q, Feng Q, Lu H, et al. Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice[J]. Nat Genet, 2018, 50(2):278-284.
doi: 10.1038/s41588-018-0041-z URL |
| [67] |
Jayakodi M, Padmarasu S, Haberer G, et al. The barley Pan-genome reveals the hidden legacy of mutation breeding[J]. Nature, 2020, 588(7837):284-289.
doi: 10.1038/s41586-020-2947-8 URL |
| [68] |
Walkowiak S, Gao LL, Monat C, et al. Multiple wheat genomes reveal global variation in modern breeding[J]. Nature, 2020. 588(7837):277-283
doi: 10.1038/s41586-020-2961-x URL |
| [69] |
Eizenga JM, Novak AM, Sibbesen JA, et al. Pangenome graphs[J]. Annu Rev Genom Hum Genet, 2020, 21(1):139-162.
doi: 10.1146/annurev-genom-120219-080406 URL |
| [70] |
Li H, Feng X, Chu C. The design and construction of reference pangenome graphs with minigraph[J]. Genome Biol, 2020, 21(1):265.
doi: 10.1186/s13059-020-02168-z URL |
| [71] |
Bayer PE, Golicz AA, Scheben A, et al. Plant Pan-genomes are the new reference[J]. Nat Plants, 2020, 6(8):914-920.
doi: 10.1038/s41477-020-0733-0 URL |
| [72] |
Liu Y, Du H, Li P, et al. Pan-genome of wild and cultivated soybeans[J]. Cell, 2020, 182(1):162-176.e13.
doi: 10.1016/j.cell.2020.05.023 URL |
| [73] |
祝光涛, 黄三文. 360度群体遗传变异扫描——大豆泛基因组研究[J]. 植物学报, 2020, 55(4):403-406.
doi: 10.11983/CBB20096 |
| Zhu GT, Huang SW. A 360-degree scanning of population genetic variations—a Pan-genome study of soybean[J]. Chin Bull Bot, 2020, 55(4):403-406. | |
| [74] |
Jain M, Koren S, Miga KH, et al. Nanopore sequencing and assembly of a human genome with ultra-long reads[J]. Nat Biotechnol, 2018, 36(4):338-345.
doi: 10.1038/nbt.4060 URL |
| [75] |
Lang DD, Zhang SL, Ren PP, et al. Comparison of the two up-to-date sequencing technologies for genome assembly:HiFi reads of Pacific Biosciences Sequel II system and ultralong reads of Oxford Nanopore[J]. Gigascience, 2020, 9(12):giaa123.
doi: 10.1093/gigascience/giaa123 URL |
| [1] | ZHANG Lu-yang, HAN Wen-long, XU Xiao-wen, YAO Jian, LI Fang-fang, TIAN Xiao-yuan, ZHANG Zhi-qiang. Identification and Expression Analysis of the Tobacco TCP Gene Family [J]. Biotechnology Bulletin, 2023, 39(6): 248-258. |
| [2] | LI Jing-rui, WANG Yu-bo, XIE Zi-wei, LI Chang, WU Xiao-lei, GONG Bin-bin, GAO Hong-bo. Identification and Expression Analysis of PIN Gene Family in Melon Under High Temperature Stress [J]. Biotechnology Bulletin, 2023, 39(5): 192-204. |
| [3] | GUO San-bao, SONG Mei-ling, LI Ling-xin, YAO Zi-zhao, GUI Ming-ming, HUANG Sheng-he. Cloning and Analysis of Chalcone Synthase Gene and Its Promoter from Euphorbia maculata [J]. Biotechnology Bulletin, 2023, 39(4): 148-156. |
| [4] | WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(4): 246-258. |
| [5] | YANG Lan, ZHANG Chen-xi, FAN Xue-wei, WANG Yang-guang, WANG Chun-xiu, LI Wen-ting. Gene Cloning, Expression Pattern, and Promoter Activity Analysis of Chicken BMP15 [J]. Biotechnology Bulletin, 2023, 39(4): 304-312. |
| [6] | CHEN Qiang, ZHOU Ming-kang, SONG Jia-min, ZHANG Chong, WU Long-kun. Identification and Analysis of LBD Gene Family and Expression Analysis of Fruit Development in Cucumis melo [J]. Biotechnology Bulletin, 2023, 39(3): 176-183. |
| [7] | PING Huai-lei, GUO Xue, YU Xiao, SONG Jing, DU Chun, WANG Juan, ZHANG Huai-bi. Cloning and Expression of PdANS in Paeonia delavayi and Correlation with Anthocyanin Content [J]. Biotechnology Bulletin, 2023, 39(3): 206-217. |
| [8] | SUN Hai-hang, GUAN Hui-lin, WANG Xu, WANG Tong, LI Hong-lin, PENG Wen-jie, LIU Bo-zhen, FAN Fang-ling. Effects of Biochar on the Soil Properties and Fungal Community Structure under Continuous Cropping of Panax notoginseng [J]. Biotechnology Bulletin, 2023, 39(2): 221-231. |
| [9] | XING Yuan, SONG Jian, LI Jun-yi, ZHENG Ting-ting, LIU Si-chen, QIAO Zhi-jun. Identification of AP Gene Family and Its Response Analysis to Abiotic Stress in Setaria italica [J]. Biotechnology Bulletin, 2023, 39(11): 238-251. |
| [10] | CHEN Chu-yi, YANG Xiao-mei, CHEN Sheng-yan, CHEN Bin, YUE Li-ran. Expression Analysis of the ZF-HD Gene Family in Chrysanthemum nankingense Under Drought and ABA Treatment [J]. Biotechnology Bulletin, 2023, 39(11): 270-282. |
| [11] | YANG Min, LONG Yu-qing, ZENG Juan, ZENG Mei, ZHOU Xin-ru, WANG Ling, FU Xue-sen, ZHOU Ri-bao, LIU Xiang-dan. Cloning and Function Analysis of Gene UGTPg17 and UGTPg36 in Lonicera macranthoides [J]. Biotechnology Bulletin, 2023, 39(10): 256-267. |
| [12] | GUO Zhi-hao, JIN Ze-xin, LIU Qi, GAO Li. Bioinformatics Analysis, Subcellular Localization and Toxicity Verification of Effector g11335 in Tilletia contraversa Kühn [J]. Biotechnology Bulletin, 2022, 38(8): 110-117. |
| [13] | CHEN Tian-ci, WU Shao-lan, YANG Guo-hui, JIANG Dan-xia, JIANG Yu-ji, CHEN Bing-zhi. Effects of Ganoderma resinaceum Alcohol Extract on Sleep and Intestinal Microbiota in Mice [J]. Biotechnology Bulletin, 2022, 38(8): 225-232. |
| [14] | YU Qiu-lin, MA Jing-yi, ZHAO Pan, SUN Peng-fang, HE Yu-mei, LIU Shi-biao, GUO Hui-hong. Cloning and Functional Analysis of Gynostemma pentaphyllum GpMIR156a and GpMIR166b [J]. Biotechnology Bulletin, 2022, 38(7): 186-193. |
| [15] | CHEN Jia-min, LIU Yong-jie, MA Jin-xiu, LI Dan, GONG Jie, ZHAO Chang-ping, GENG Hong-wei, GAO Shi-qing. Expression Pattern Analysis of Histone Methyltransferase Under Drought Stress in Hybrid Wheat [J]. Biotechnology Bulletin, 2022, 38(7): 51-61. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
