








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (1): 15-32.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1562
Previous Articles Next Articles
YIN Guo-liang1,2( ), SUN Wen-hao1,2, PANG Xiao-yun1, SUN Fei1,2(
), SUN Wen-hao1,2, PANG Xiao-yun1, SUN Fei1,2( )
)
Received:2021-12-16
Online:2022-01-26
Published:2022-02-22
Contact:
SUN Fei
E-mail:yinguoliang19@mails.ucas.ac.cn;feisun@ibp.ac.cn
YIN Guo-liang, SUN Wen-hao, PANG Xiao-yun, SUN Fei. Application of cryo-Electron Microscopy in Molecular Botany Research[J]. Biotechnology Bulletin, 2022, 38(1): 15-32.
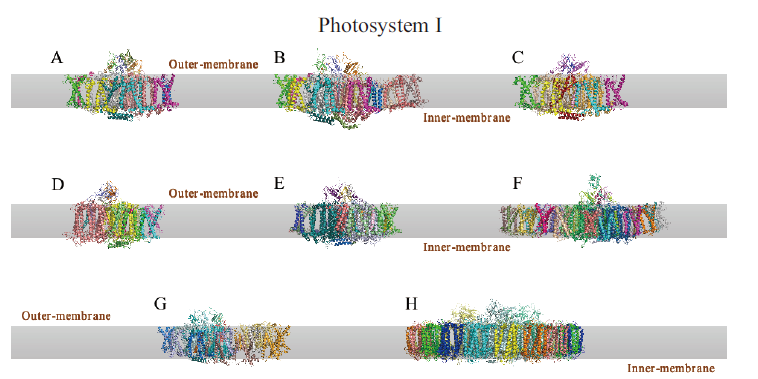
Fig. 2 cryo-EM structure of photosystem I A:PSI-FD complex of Pisum sativum;B:PSI-LHCI-LHCII complex of Zea mays;C:PSI complex of Dunaliella salina;D:PSI complex of Cyanidioschyzon merolae;E:PSI complex of Chlamydomonas reinhardtii;F:PSI-FCP complex of Chaeto-ceros gracilis;G:PSI complex of Physcomitrium patens;H:PSI-isiA complex of Thermosynechococcus vulcanus
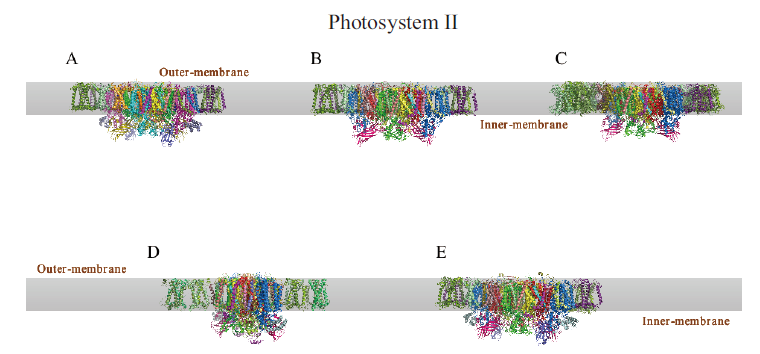
Fig.3 Cryo-EM structure of photosystem II A:PSII-LHCII complex of Spinacia oleracea. B:PSII-LHCII complex of Pisum sativum. C:PSII complex of Arabidopsis thaliana. D:C2S2 type PSII-FCPII complex of Chaetoceros gracilis. E:C2S2 type PSII-LHCII complex of Chlamydomonas reinhardtii

Fig. 9 cryo-EM structure and activated conformation of OSCA1 Cryo-EM structure of AtOSCA1.1(A),AtOSCA3.1(B);(C)Conformational changes of AtOSCA1.1 to allow Ca2 + entry
| Name | Organism(s) | Resolution | Year | PDB | Reference | |
|---|---|---|---|---|---|---|
| Photosynthesis | PSII-LHCII supercomplex | Spinacia oleracea | 3.20 Å | 2016 | 3JCU | [ |
| M-LHCII and CP24 complexes | Pisum sativum | 3.50 Å | 2017 | 5XNO | [ | |
| C2S2M2N2-type PSII-LHCII | Pisum sativum | 2.70 Å | 2017 | 5XNL | [ | |
| Phycobilisome | Griffithsia pacifica | 3.50 Å | 2017 | 5Y6P | [ | |
| PSI-LHCR | Cyanidioschyzon merolae strain 10D | 3.82 Å | 2018 | 5ZGH | [ | |
| Photosystem I supercomplex with light-harvesting complexes I and II | Zea mays,Zea mays subsp. mays | 3.30 Å | 2018 | 5ZJI | [ | |
| PSII-FCP supercomplex | Chaetoceros gracilis | 3.02 Å | 2019 | 6JLU | [ | |
| C2S2M2L2-type PSII-LHCII supercomplex | Chlamydomonas reinhardtii | 3.40 Å | 2019 | 6KAD | [ | |
| C2S2-type PSII-LHCII supercomplex | Chlamydomonas reinhardtii | 2.70 Å | 2019 | 6KAC | [ | |
| Photosystem I | Chlamydomonas reinhardtii | 3.30 Å | 2019 | 6IJO | [ | |
| Cytochrome b6f complex | Spinacia oleracea | 3.58 Å | 2019 | 6RQF | [ | |
| Phycobilisome | Porphyridium purpureum | 2.80 Å | 2019 | 6KGX | [ | |
| NDH | Thermosynechococcus vestitus BP-1 | 3.10 Å | 2019 | 6NBY | [ | |
| Photosystem I | Dunaliella salina | 3.20 Å | 2020 | 6RHZ | [ | |
| PSI-FCPI supercomplex | Chaetoceros gracilis | 2.40 Å | 2020 | 6L4U | [ | |
| Photosystem I complex | Synechocystis sp. PCC 6803 substr. Kazusa | 3.10 Å | 2020 | 6UZV | [ | |
| Fd-NDH-1L complex | Thermosynechococcus elongatus BP-1 | 3.20 Å | 2020 | 6L7O | [ | |
| NDH-1LdelV complex | Thermosynechococcus elongatus BP-1 | 3.60 Å | 2020 | 6L7P | [ | |
| PSI-NDH supercomplex | Hordeum vulgare subsp. spontaneum | 4.50 Å | 2021 | 7F9O | [ | |
| Chloroplast NDH complex | Hordeum vulgare subsp. spontaneum | 3.70 Å | 2021 | 7EU3 | [ | |
| PSI-LHCI-Lhca5 supercomplex | Hordeum vulgare subsp. spontaneum | 3.40 Å | 2021 | 7EW6 | [ | |
| PSI-LHCI-Lhca6 supercomplex | Hordeum vulgare subsp. spontaneum | 3.88 Å | 2021 | 7EWK | [ | |
| Immunity | NLR complex | Arabidopsis thaliana | 3.70 Å | 2019 | 6J5W | [ |
| NLR RPP1 LRR-ID domain in complex with ATR1 | Hyaloperonospora arabidopsidis Emoy2,Arabidopsis thaliana | 3.16 Å | 2020 | 7CRB | [ | |
| Activated Roq1 resistosome | Nicotiana benthamiana,Xanthomonas euvesicatoria | 3.80 Å | 2020 | 7JLU | [ | |
| Transportation | atOSCA3.1 channel | Arabidopsis thaliana | 4.80 Å | 2018 | 5Z1F | [ |
| atOSCA1.1 channel | Arabidopsis thaliana | 3.52 Å | 2018 | 6JPF | [ | |
| Activated ion channel OSCA1.2 | Arabidopsis thaliana | 3.10 Å | 2018 | 6MGV | [ | |
| Cation channel | Arabidopsis thaliana | 3.68 Å | 2018 | 6IJZ | [ | |
| Ion channel OSCA1.2 | Oryza sativa | 4.90 Å | 2019 | 6OCE | [ | |
| MSL1 | Arabidopsis thaliana | 3.06 Å | 2020 | 6VXM | [ |
Table 1 Structure of important plant-related biological macromolecules solved by cryo-EM
| Name | Organism(s) | Resolution | Year | PDB | Reference | |
|---|---|---|---|---|---|---|
| Photosynthesis | PSII-LHCII supercomplex | Spinacia oleracea | 3.20 Å | 2016 | 3JCU | [ |
| M-LHCII and CP24 complexes | Pisum sativum | 3.50 Å | 2017 | 5XNO | [ | |
| C2S2M2N2-type PSII-LHCII | Pisum sativum | 2.70 Å | 2017 | 5XNL | [ | |
| Phycobilisome | Griffithsia pacifica | 3.50 Å | 2017 | 5Y6P | [ | |
| PSI-LHCR | Cyanidioschyzon merolae strain 10D | 3.82 Å | 2018 | 5ZGH | [ | |
| Photosystem I supercomplex with light-harvesting complexes I and II | Zea mays,Zea mays subsp. mays | 3.30 Å | 2018 | 5ZJI | [ | |
| PSII-FCP supercomplex | Chaetoceros gracilis | 3.02 Å | 2019 | 6JLU | [ | |
| C2S2M2L2-type PSII-LHCII supercomplex | Chlamydomonas reinhardtii | 3.40 Å | 2019 | 6KAD | [ | |
| C2S2-type PSII-LHCII supercomplex | Chlamydomonas reinhardtii | 2.70 Å | 2019 | 6KAC | [ | |
| Photosystem I | Chlamydomonas reinhardtii | 3.30 Å | 2019 | 6IJO | [ | |
| Cytochrome b6f complex | Spinacia oleracea | 3.58 Å | 2019 | 6RQF | [ | |
| Phycobilisome | Porphyridium purpureum | 2.80 Å | 2019 | 6KGX | [ | |
| NDH | Thermosynechococcus vestitus BP-1 | 3.10 Å | 2019 | 6NBY | [ | |
| Photosystem I | Dunaliella salina | 3.20 Å | 2020 | 6RHZ | [ | |
| PSI-FCPI supercomplex | Chaetoceros gracilis | 2.40 Å | 2020 | 6L4U | [ | |
| Photosystem I complex | Synechocystis sp. PCC 6803 substr. Kazusa | 3.10 Å | 2020 | 6UZV | [ | |
| Fd-NDH-1L complex | Thermosynechococcus elongatus BP-1 | 3.20 Å | 2020 | 6L7O | [ | |
| NDH-1LdelV complex | Thermosynechococcus elongatus BP-1 | 3.60 Å | 2020 | 6L7P | [ | |
| PSI-NDH supercomplex | Hordeum vulgare subsp. spontaneum | 4.50 Å | 2021 | 7F9O | [ | |
| Chloroplast NDH complex | Hordeum vulgare subsp. spontaneum | 3.70 Å | 2021 | 7EU3 | [ | |
| PSI-LHCI-Lhca5 supercomplex | Hordeum vulgare subsp. spontaneum | 3.40 Å | 2021 | 7EW6 | [ | |
| PSI-LHCI-Lhca6 supercomplex | Hordeum vulgare subsp. spontaneum | 3.88 Å | 2021 | 7EWK | [ | |
| Immunity | NLR complex | Arabidopsis thaliana | 3.70 Å | 2019 | 6J5W | [ |
| NLR RPP1 LRR-ID domain in complex with ATR1 | Hyaloperonospora arabidopsidis Emoy2,Arabidopsis thaliana | 3.16 Å | 2020 | 7CRB | [ | |
| Activated Roq1 resistosome | Nicotiana benthamiana,Xanthomonas euvesicatoria | 3.80 Å | 2020 | 7JLU | [ | |
| Transportation | atOSCA3.1 channel | Arabidopsis thaliana | 4.80 Å | 2018 | 5Z1F | [ |
| atOSCA1.1 channel | Arabidopsis thaliana | 3.52 Å | 2018 | 6JPF | [ | |
| Activated ion channel OSCA1.2 | Arabidopsis thaliana | 3.10 Å | 2018 | 6MGV | [ | |
| Cation channel | Arabidopsis thaliana | 3.68 Å | 2018 | 6IJZ | [ | |
| Ion channel OSCA1.2 | Oryza sativa | 4.90 Å | 2019 | 6OCE | [ | |
| MSL1 | Arabidopsis thaliana | 3.06 Å | 2020 | 6VXM | [ |
| [1] |
Henderson R, Unwin PN. Three-dimensional model of purple membrane obtained by electron microscopy[J]. Nature, 1975, 257(5521):28-32.
doi: 10.1038/257028a0 URL |
| [2] |
Unwin PN, Henderson R. Molecular structure determination by electron microscopy of unstained crystalline specimens[J]. J Mol Biol, 1975, 94(3):425-440.
pmid: 1236957 |
| [3] |
Henderson R, Baldwin JM, Ceska TA, et al. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy[J]. J Mol Biol, 1990, 213(4):899-929.
pmid: 2359127 |
| [4] |
Henderson R. The potential and limitations of neutrons, electrons and X-rays for atomic resolution microscopy of unstained biological molecules[J]. Q Rev Biophys, 1995, 28(2):171-193.
pmid: 7568675 |
| [5] |
Adrian M, Dubochet J, Lepault J, et al. Cryo-electron microscopy of viruses[J]. Nature, 1984, 308(5954):32-36.
doi: 10.1038/308032a0 URL |
| [6] |
Downing KH. Observations of restricted beam-induced specimen motion with small-spot illumination[J]. Ultramicroscopy, 1988, 24(4):387-397.
pmid: 3363743 |
| [7] |
Hayward SB, Glaeser RM. Radiation damage of purple membrane at low temperature[J]. Ultramicroscopy, 1979, 04(2):201-210.
pmid: 473421 |
| [8] |
Frank J. Classification of macromolecular assemblies studied as ‘single particles’[J]. Q Rev Biophys, 1990, 23(3):281-329.
pmid: 2204955 |
| [9] |
Russo CJ, Passmore LA. Controlling protein adsorption on graphene for cryo-EM using low-energy hydrogen plasmas[J]. Nat Methods, 2014, 11(6):649-652.
doi: 10.1038/NMETH.2931 |
| [10] |
Pantelic RS, Meyer JC, Kaiser U, et al. Graphene oxide:a substrate for optimizing preparations of frozen-hydrated samples[J]. J Struct Biol, 2010, 170(1):152-156.
doi: 10.1016/j.jsb.2009.12.020 pmid: 20035878 |
| [11] |
Russo CJ, Passmore LA. Electron microscopy:Ultrastable gold substrates for electron cryomicroscopy[J]. Science, 2014, 346(6215):1377-1380.
doi: 10.1126/science.1259530 URL |
| [12] |
Huang XJ, Zhang L, Wen ZL, et al. Amorphous nickel titanium alloy film:a new choice for cryo electron microscopy sample preparation[J]. Prog Biophys Mol Biol, 2020, 156:3-13.
doi: 10.1016/j.pbiomolbio.2020.07.009 URL |
| [13] |
Fan HC, Wang B, Zhang Y, et al. A cryo-electron microscopy support film formed by 2D crystals of hydrophobin HFBI[J]. Nat Commun, 2021, 12(1):7257.
doi: 10.1038/s41467-021-27596-8 URL |
| [14] |
Ravelli RBG, Nijpels FJT, Henderikx RJM, et al. Cryo-EM structures from sub-nl volumes using pin-printing and jet vitrification[J]. Nat Commun, 2020, 11(1):2563.
doi: 10.1038/s41467-020-16392-5 URL |
| [15] |
Noble AJ, Wei H, Dandey VP, et al. Reducing effects of particle adsorption to the air-water interface in cryo-EM[J]. Nat Methods, 2018, 15(10):793-795.
doi: 10.1038/s41592-018-0139-3 pmid: 30250056 |
| [16] |
Bharat TAM, Scheres SHW. Resolving macromolecular structures from electron cryo-tomography data using subtomogram averaging in RELION[J]. Nat Protoc, 2016, 11(11):2054-2065.
doi: 10.1038/nprot.2016.124 URL |
| [17] |
Punjani A, Rubinstein JL, Fleet DJ, et al. cryoSPARC:algorithms for rapid unsupervised cryo-EM structure determination[J]. Nat Methods, 2017, 14(3):290-296.
doi: 10.1038/nmeth.4169 pmid: 28165473 |
| [18] |
Tegunov D, Cramer P. Real-time cryo-electron microscopy data preprocessing with warp[J]. Nat Methods, 2019, 16(11):1146-1152.
doi: 10.1038/s41592-019-0580-y URL |
| [19] |
Wu CL, Huang XJ, Cheng J, et al. High-quality, high-throughput cryo-electron microscopy data collection via beam tilt and astigmatism-free beam-image shift[J]. J Struct Biol, 2019, 208(3):107396.
doi: 10.1016/j.jsb.2019.09.013 URL |
| [20] |
Liao MF, Cao EH, Julius D, et al. Structure of the TRPV1 ion channel determined by electron cryo-microscopy[J]. Nature, 2013, 504(7478):107-112.
doi: 10.1038/nature12822 URL |
| [21] |
Yip KM, Fischer N, Paknia E, et al. Atomic-resolution protein structure determination by cryo-EM[J]. Nature, 2020, 587(7832):157-161.
doi: 10.1038/s41586-020-2833-4 URL |
| [22] |
Nakane T, Kotecha A, Sente A, et al. Single-particle cryo-EM at atomic resolution[J]. Nature, 2020, 587(7832):152-156.
doi: 10.1038/s41586-020-2829-0 URL |
| [23] | Wan RX, Bai R, Yan CY, et al. Structures of the catalytically activated yeast spliceosome reveal the mechanism of branching[J]. Cell, 2019, 177(2):339-351. e13. |
| [24] |
Wang N, Zhao DM, Wang JL, et al. Architecture of African swine fever virus and implications for viral assembly[J]. Science, 2019, 366(6465):640-644.
doi: 10.1126/science.aaz1439 pmid: 31624094 |
| [25] |
Andrés G, Charro D, Matamoros T, et al. The cryo-EM structure of African swine fever virus unravels a unique architecture comprising two icosahedral protein capsids and two lipoprotein membranes[J]. J Biol Chem, 2020, 295(1):1-12.
doi: 10.1074/jbc.AC119.011196 URL |
| [26] |
Liang YL, Khoshouei M, Radjainia M, et al. Phase-plate cryo-EM structure of a class B GPCR-G-protein complex[J]. Nature, 2017, 546(7656):118-123.
doi: 10.1038/nature22327 URL |
| [27] | de Waal P, Zhou XE, He YZ, et al. Structural identification of phosphorylation codes for arrestin recruitment by G protein-coupled receptors[J]. Protein Sci, 2017, 26:133-133. |
| [28] |
Du J, Wang DJ, Fan HC, et al. Structures of human mGlu2 and mGlu7 homo- and heterodimers[J]. Nature, 2021, 594(7864):589-593.
doi: 10.1038/s41586-021-03641-w URL |
| [29] |
Kang YY, Kuybeda O, de Waal PW, et al. Cryo-EM structure of human rhodopsin bound to an inhibitory G protein[J]. Nature, 2018, 558(7711):553-558.
doi: 10.1038/s41586-018-0215-y URL |
| [30] |
Liang YL, Khoshouei M, Glukhova A, et al. Phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex[J]. Nature, 2018, 555(7694):121-125.
doi: 10.1038/nature25773 URL |
| [31] |
Qiao AN, Han S, Li XM, et al. Structural basis of G s and G i recognition by the human glucagon receptor[J]. Science, 2020, 367(6484):1346-1352.
doi: 10.1126/science.aaz5346 URL |
| [32] |
Zhang Y, Sun BF, Feng D, et al. Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein[J]. Nature, 2017, 546(7657):248-253.
doi: 10.1038/nature22394 URL |
| [33] | Zhou XE, He YZ, de Waal PW, et al. Identification of phosphorylation codes for arrestin recruitment by G protein-coupled receptors[J]. Cell, 2017, 170(3):457-469. e13. |
| [34] | Gong HR, Li J, Xu A, et al. An electron transfer path connects subunits of a mycobacterial respiratory super complex[J]. Science, 2018, 362(6418):eaat8923. |
| [35] |
Zong S, Wu M, Gu JK, et al. Structure of the intact 14-subunit human cytochrome c oxidase[J]. Cell Res, 2018, 28(10):1026-1034.
doi: 10.1038/s41422-018-0071-1 URL |
| [36] |
Zhu GL, Zeng H, Zhang SB, et al. A 3. 3 Å-resolution structure of hyperthermophilic respiratory complex III reveals the mechanism of its thermal stability[J]. Angew Chem Int Ed Engl, 2020, 59(1):343-351.
doi: 10.1002/anie.v59.1 URL |
| [37] |
Wrapp D, Wang NS, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation[J]. Science, 2020, 367(6483):1260-1263.
doi: 10.1126/science.abb2507 URL |
| [38] |
Turo\u0148ová B, Sikora M, Schürmann C, et al. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges[J]. Science, 2020, 370(6513):203-208.
doi: 10.1126/science.abd5223 URL |
| [39] | Long SW, Olsen RJ, Christensen PA, et al. Molecular architecture of early dissemination and massive second wave of the SARS-CoV-2 virus in a major metropolitan area[J]. medRxiv, 2020:2020 Sep 29;2020. 09. 22. 20199125. |
| [40] | Yao HP, Song YT, Chen Y, et al. Molecular architecture of the SARS-CoV-2 virus[J]. Cell, 2020, 183(3):730-738. e13. |
| [41] | Walls AC, Park YJ, Tortorici MA, et al. Structure, function, antigenicity of the SARS-CoV-2 spike glycoprotein[J]. Cell, 2020, 181(2):281-292. e6. |
| [42] |
Ke ZL, Oton J, Qu K, et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions[J]. Nature, 2020, 588(7838):498-502.
doi: 10.1038/s41586-020-2665-2 URL |
| [43] | Wang Q, Wu JQ, Wang HF, et al. Structural basis for RNA replication by the SARS-CoV-2 polymerase[J]. Cell, 2020, 182(2):417-428. e13. |
| [44] |
Lucić V, Förster F, Baumeister W. Structural studies by electron tomography:from cells to molecules[J]. Annu Rev Biochem, 2005, 74:833-865.
doi: 10.1146/biochem.2005.74.issue-1 URL |
| [45] |
Danev R, Baumeister W. Expanding the boundaries of cryo-EM with phase plates[J]. Curr Opin Struct Biol, 2017, 46:87-94.
doi: 10.1016/j.sbi.2017.06.006 URL |
| [46] |
Wang HW, Fan X. Challenges and opportunities in cryo-EM with phase plate[J]. Curr Opin Struct Biol, 2019, 58:175-182.
doi: 10.1016/j.sbi.2019.06.013 URL |
| [47] | Bieber A, Capitanio C, Wilfling F, et al. Sample preparation by 3D-correlative focused ion beam milling for high-resolution cryo-electron tomography[J]. J Vis Exp, 2021(176):e62886. |
| [48] | Moravcová J, Pinkas M, Holbová R, et al. Preparation and cryo-FIB micromachining of Saccharomyces cerevisiae for cryo-electron tomography[J]. J Vis Exp, 2021(177):e62351. |
| [49] |
Zhang JG, Zhang DY, Sun L, et al. VHUT-cryo-FIB, a method to fabricate frozen hydrated lamellae from tissue specimens for in situ cryo-electron tomography[J]. J Struct Biol, 2021, 213(3):107763.
doi: 10.1016/j.jsb.2021.107763 URL |
| [50] | Hu H, Santiveri M, Wadhwa N, et al. Structural basis of torque generation in the bi-directional bacterial flagellar motor[J]. Trends Biochem Sci, 2021, S0968-0004(21)00139-0. |
| [51] |
Namba K. A proposed gear mechanism for torque generation in the flagellar motor[J]. Nat Struct Mol Biol, 2020, 27(11):1004-1006.
doi: 10.1038/s41594-020-00514-0 URL |
| [52] |
Zhao XW, Norris SJ, Liu J. Molecular architecture of the bacterial flagellar motor in cells[J]. Biochemistry, 2014, 53(27):4323-4333.
doi: 10.1021/bi500059y URL |
| [53] | Zhu SW, Qin Z, Wang JY, et al. In situ structural analysis of the spirochetal flagellar motor by cryo-electron tomography[J]. Methods Mol Biol, 2017, 1593:229-242. |
| [54] |
Dimchev G, Amiri B, Fäβler F, et al. Computational toolbox for ultrastructural quantitative analysis of filament networks in cryo-ET data[J]. J Struct Biol, 2021, 213(4):107808.
doi: 10.1016/j.jsb.2021.107808 pmid: 34742832 |
| [55] |
Sazzed S, Song JH, Kovacs JA, et al. Tracing actin filament bundles in three-dimensional electron tomography density maps of hair cell stereocilia[J]. Molecules, 2018, 23(4):882.
doi: 10.3390/molecules23040882 URL |
| [56] | Kittisopikul M, Shimi T, Tatli M, et al. Computational analyses reveal spatial relationships between nuclear pore complexes and specific lamins[J]. J Cell Biol, 2021, 220(4):e202007082. |
| [57] |
Mahamid J, Pfeffer S, Schaffer M, et al. Visualizing the molecular sociology at the HeLa cell nuclear periphery[J]. Science, 2016, 351(6276):969-972.
doi: 10.1126/science.aad8857 pmid: 26917770 |
| [58] |
Zhang YQ, Li S, Zeng C, et al. Molecular architecture of the luminal ring of the Xenopus laevis nuclear pore complex[J]. Cell Res, 2020, 30(6):532-540.
doi: 10.1038/s41422-020-0320-y URL |
| [59] | 孙飞. 生物三维电子显微成像技术展望[M]. 中国科学院创新发展研究中心. 中国生命健康2035-技术预见. 北京: 科学出版社, 2019:241-256. |
| Sun Fei. Prospect of biological three-dimensional electron microscopy imaging technology[M]. Center for Innovation and Development Chinese Academy of Sciences. Technology Foresight Towards 2035 in China: Life and Health. Science Press. 2019:241-256. | |
| [60] |
Anderson S, Anderson HL, Bashall A, et al. Assembly and crystal structure of a photoactive array of five porphyrins[J]. Angew Chem Int Ed Engl, 1995, 34(10):1096-1099.
doi: 10.1002/(ISSN)1521-3773 URL |
| [61] |
Matsumura H, Xie Y, Shirakata S, et al. Crystal structures of C4 form maize and quaternary complex of E. coli phosphoenolpyruvate carboxylases[J]. Structure, 2002, 10(12):1721-1730.
doi: 10.1016/S0969-2126(02)00913-9 URL |
| [62] |
Toporik H, Khmelnitskiy A, Dobson Z, et al. The structure of a red-shifted photosystem I reveals a red site in the core antenna[J]. Nat Commun, 2020, 11:5279.
doi: 10.1038/s41467-020-18884-w URL |
| [63] |
Pi X, Tian LR, Dai HE, et al. Unique organization of photosystem I-light-harvesting super complex revealed by cryo-EM from a red alga[J]. Proc Natl Acad Sci USA, 2018, 115(17):4423-4428.
doi: 10.1073/pnas.1722482115 URL |
| [64] |
Kubota-Kawai H, Burton-Smith RN, Tokutsu R, et al. Ten antenna proteins are associated with the core in the supramolecular organization of the photosystem I super complex in Chlamydomonas reinhardtii[J]. J Biol Chem, 2019, 294(12):4304-4314.
doi: 10.1074/jbc.RA118.006536 pmid: 30670590 |
| [65] |
Su XD, Ma J, Pan XW, et al. Antenna arrangement and energy transfer pathways of a green algal photosystem-I-LHCI super complex[J]. Nat Plants, 2019, 5(3):273-281.
doi: 10.1038/s41477-019-0380-5 URL |
| [66] |
Perez-Boerema A, Klaiman D, Caspy I, et al. Structure of a minimal photosystem I from the green alga Dunaliella salina[J]. Nat Plants, 2020, 6(3):321-327.
doi: 10.1038/s41477-020-0611-9 pmid: 32123351 |
| [67] |
Nagao R, Kato K, Ifuku K, et al. Structural basis for assembly and function of a diatom photosystem I-light-harvesting super complex[J]. Nat Commun, 2020, 11(1):2481.
doi: 10.1038/s41467-020-16324-3 pmid: 32424145 |
| [68] |
Pan XW, Ma J, Su XD, et al. Structure of the maize photosystem I super complex with light-harvesting complexes I and II[J]. Science, 2018, 360(6393):1109-1113.
doi: 10.1126/science.aat1156 URL |
| [69] | Shen L, Tang K, Wang W, et al. Architecture of the chloroplast PSI-NDH super complex in Hordeum vulgare[J]. Nature, 2021: 2021 Dec 8. |
| [70] |
Zhang CL, Shuai J, Ran ZX, et al. Structural insights into NDH-1 mediated cyclic electron transfer[J]. Nat Commun, 2020, 11(1):888.
doi: 10.1038/s41467-020-14732-z URL |
| [71] |
Wei XP, Su XD, Cao P, et al. Structure of spinach photosystem II-LHCII super complex at 3. 2 Å resolution[J]. Nature, 2016, 534(7605):69-74.
doi: 10.1038/nature18020 URL |
| [72] |
Su XD, Ma J, Wei XP, et al. Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII super complex[J]. Science, 2017, 357(6353):815-820.
doi: 10.1126/science.aan0327 URL |
| [73] |
van Bezouwen LS, Caffarri S, Kale RS, et al. Subunit and chlorophyll organization of the plant photosystem II super complex[J]. Nat Plants, 2017, 3:17080.
doi: 10.1038/nplants.2017.80 pmid: 28604725 |
| [74] | Pi X, Zhao SH, Wang WD, et al. The pigment-protein network of a diatom photosystem II-light-harvesting antenna super complex[J]. Science, 2019, 365(6452):eaax4406. |
| [75] |
Sheng X, Watanabe A, Li AJ, et al. Structural insight into light harvesting for photosystem II in green algae[J]. Nat Plants, 2019, 5(12):1320-1330.
doi: 10.1038/s41477-019-0543-4 pmid: 31768031 |
| [76] |
Malone LA, Qian P, Mayneord GE, et al. Cryo-EM structure of the spinach cytochrome b 6 f complex at 3. 6 Å resolution[J]. Nature, 2019, 575(7783):535-539.
doi: 10.1038/s41586-019-1746-6 URL |
| [77] |
Laughlin TG, Bayne AN, Trempe JF, et al. Structure of the complex I-like molecule NDH of oxygenic photosynjournal[J]. Nature, 2019, 566(7744):411-414.
doi: 10.1038/s41586-019-0921-0 URL |
| [78] |
Zhang J, Ma JF, Liu DS, et al. Structure of phycobilisome from the red alga Griffithsia Pacifica[J]. Nature, 2017, 551(7678):57-63.
doi: 10.1038/nature24278 URL |
| [79] |
Ma JF, You X, Sun S, et al. Structural basis of energy transfer in Porphyridium purpureum phycobilisome[J]. Nature, 2020, 579(7797):146-151.
doi: 10.1038/s41586-020-2020-7 URL |
| [80] |
Li MJ, Ma JF, Li XM, et al. In situ cryo-ET structure of phycobilisome-photosystem II super complex from red alga[J]. eLife, 2021, 10:e69635.
doi: 10.7554/eLife.69635 URL |
| [81] |
Zhu JK. Abiotic stress signaling and responses in plants[J]. Cell, 2016, 167(2):313-324.
doi: 10.1016/j.cell.2016.08.029 URL |
| [82] |
Song W, Forderer A, Yu DL, et al. Structural biology of plant defence[J]. New Phytol, 2021, 229(2):692-711.
doi: 10.1111/nph.v229.2 URL |
| [83] |
Wang W, Feng BM, Zhou JM, et al. Plant immune signaling:advancing on two frontiers[J]. J Integr Plant Biol, 2020, 62(1):2-24.
doi: 10.1111/jipb.v62.1 URL |
| [84] |
Wang JZ, Chai JJ. Structural insights into the plant immune receptors PRRs and NLRs[J]. Plant Physiol, 2020, 182(4):1566-1581.
doi: 10.1104/pp.19.01252 URL |
| [85] | 赵燕波, 郭婉兰, 张丹丹, 等. 植物免疫受体病原体识别机制的结构生物学研究进展[J]. 福建师范大学学报:自然科学版, 2020, 36(2):12-24. |
| Zhao YB, Guo WL, Zhang DD, et al. Progress in structural basis for pathogen recognition mechanism of plant immune receptors[J]. J Fujian Norm Univ Nat Sci Ed, 2020, 36(2):12-24. | |
| [86] | 邓一文, 刘裕强, 王静, 等. 农作物抗病虫研究的战略思考[J]. 中国科学:生命科学, 2021, 51:1435-1446. |
| Deng YW, Liu YQ, Wang J, et al. Strategic thinking and research on crop disease and pest resistance in China[J]. Sci Sin Vitae, 2021, 51:1435-1446. | |
| [87] |
Jones JDG, Dangl JL. The plant immune system[J]. Nature, 2006, 444(7117):323-329.
doi: 10.1038/nature05286 URL |
| [88] | Jones JDG, Vance RE, Dangl JL. Intracellular innate immune surveillance devices in plants and animals[J]. Science, 2016, 354(6316):aaf6395. |
| [89] | Ngou BPM, Ahn HK, Ding PT, et al. Mutual potentiation of plant immunity by cell-surface and intracellular receptors[J]. Nature, 2021, 592(7852):110-115. |
| [90] | Yuan MH, Jiang ZY, Bi GZ, et al. Pattern-recognition receptors are required for NLR-mediated plant immunity[J]. Nature, 2021, 592(7852):105-109. |
| [91] |
Pruitt RN, Gust AA, Nürnberger T. Plant immunity unified[J]. Nat Plants, 2021, 7(4):382-383.
doi: 10.1038/s41477-021-00903-3 URL |
| [92] | Chang M, Chen H, Liu F, et al. PTI and ETI:convergent pathways with diverse elicitors[J]. Trends Plant Sci, 2021: 2021 Dec 1;S1360-2021 Dec 1;S1385(21)00319-8. |
| [93] |
Boutrot F, Zipfel C. Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance[J]. Annu Rev Phytopathol, 2017, 55:257-286.
doi: 10.1146/annurev-phyto-080614-120106 pmid: 28617654 |
| [94] |
Liu ZX, Wu Y, Yang F, et al. BIK1 interacts with PEPRs to mediate ethylene-induced immunity[J]. Proc Natl Acad Sci USA, 2013, 110(15):6205-6210.
doi: 10.1073/pnas.1215543110 URL |
| [95] |
Iizasa E, Mitsutomi M, Nagano Y. Direct binding of a plant LysM receptor-like kinase, LysM RLK1/CERK1, to chitin in vitro[J]. J Biol Chem, 2010, 285(5):2996-3004.
doi: 10.1074/jbc.M109.027540 URL |
| [96] |
Hayafune M, Berisio R, Marchetti R, et al. Chitin-induced activation of immune signaling by the rice receptor CEBiP relies on a unique sandwich-type dimerization[J]. Proc Natl Acad Sci USA, 2014, 111(3):E404-E413.
doi: 10.1073/pnas.1312099111 URL |
| [97] |
Wang JZ, Chai JJ. Molecular actions of NLR immune receptors in plants and animals[J]. Sci China Life Sci, 2020, 63(9):1303-1316.
doi: 10.1007/s11427-019-1687-6 URL |
| [98] | Bentham A, Burdett H, Anderson PA, et al. Animal NLRs provide structural insights into plant NLR function[J]. Ann Bot, 2017, 119(5):827-702. |
| [99] | Wang JZ, Wang J, Hu MJ, et al. Ligand-triggered allosteric ADP release primes a plant NLR complex[J]. Science, 2019, 364(6435):eaav5868. |
| [100] | Wang JZ, Hu MJ, Wang J, et al. Reconstitution and structure of a plant NLR resistosome conferring immunity[J]. Science, 2019, 364(6435):eaav5870. |
| [101] | Bi GZ, Su M, Li N, et al. The ZAR1 resistosome is a calcium-permeable channel triggering plant immune signaling[J]. Cell, 2021, 184(13):3528-3541. e12. |
| [102] | Ma SC, Lapin D, Liu L, et al. Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme[J]. Science, 2020, 370(3521):eabe3069. |
| [103] | Martin R, Qi TC, Zhang HB, et al. Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ[J]. Science, 2020, 370(3521):eabd9993. |
| [104] |
Brutus A, Sicilia F, Macone A, et al. A domain swap approach reveals a role of the plant wall-associated kinase 1(WAK1)as a receptor of oligogalacturonides[J]. Proc Natl Acad Sci USA, 2010, 107(20):9452-9457.
doi: 10.1073/pnas.1000675107 URL |
| [105] |
Choi J, Tanaka K, Cao YR, et al. Identification of a plant receptor for extracellular ATP[J]. Science, 2014, 343(6168):290-294.
doi: 10.1126/science.343.6168.290 URL |
| [106] |
Wang LM, Wilkins KA, Davies JM. Arabidopsis DORN1 extracellular ATP receptor;activation of plasma membrane K +-and Ca2 +-permeable conductances[J]. New Phytol, 2018, 218(4):1301-1304.
doi: 10.1111/nph.2018.218.issue-4 URL |
| [107] |
Gong ZZ, Xiong LM, Shi HZ, et al. Plant abiotic stress response and nutrient use efficiency[J]. Sci China Life Sci, 2020, 63(5):635-674.
doi: 10.1007/s11427-020-1683-x URL |
| [108] | 朱健康, 倪建平. 植物非生物胁迫信号转导及应答[J]. 中国稻米, 2016, 22(6):52-60. |
| Zhu JK, Ni JP. Abiotic stress signaling and responses in plants[J]. China Rice, 2016, 22(6):52-60. | |
| [109] |
Yuan F, Yang HM, Xue Y, et al. OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis[J]. Nature, 2014, 514(7522):367-371.
doi: 10.1038/nature13593 URL |
| [110] |
Basu D, Haswell ES. Plant mechanosensitive ion channels:an ocean of possibilities[J]. Curr Opin Plant Biol, 2017, 40:43-48.
doi: 10.1016/j.pbi.2017.07.002 URL |
| [111] |
Lee CP, Maksaev G, Jensen GS, et al. MSL1 is a mechanosensitive ion channel that dissipates mitochondrial membrane potential and maintains redox homeostasis in mitochondria during abiotic stress[J]. Plant J, 2016, 88(5):809-825.
doi: 10.1111/tpj.2016.88.issue-5 URL |
| [112] |
Zhang MF, Wang DL, Kang YL, et al. Structure of the mechanosensitive OSCA channels[J]. Nat Struct Mol Biol, 2018, 25(9):850-858.
doi: 10.1038/s41594-018-0117-6 URL |
| [113] |
Jojoa-Cruz S, Saotome K, Murthy SE, et al. Cryo-EM structure of the mechanically activated ion channel OSCA1. 2[J]. eLife, 2018, 7:e41845.
doi: 10.7554/eLife.41845 URL |
| [114] |
Murthy SE, Dubin AE, Whitwam T, et al. OSCA/TMEM63 are an evolutionarily conserved family of mechanically activated ion channels[J]. eLife, 2018, 7:e41844.
doi: 10.7554/eLife.41844 URL |
| [115] |
Liu X, Wang JW, Sun LF. Structure of the hyperosmolality-gated calcium-permeable channel OSCA1. 2[J]. Nat Commun, 2018, 9(1):5060.
doi: 10.1038/s41467-018-07564-5 URL |
| [116] |
Maity K, Heumann JM, McGrath AP, et al. Cryo-EM structure of OSCA1. 2 from Oryza sativa elucidates the mechanical basis of potential membrane hyperosmolality gating[J]. Proc Natl Acad Sci USA, 2019, 116(28):14309-14318.
doi: 10.1073/pnas.1900774116 URL |
| [117] | Li YW, Hu YF, Wang JW, et al. Structural insights into a plant mechanosensitive ion channel MSL1[J]. Cell Rep, 2020, 30(13):4518-4527. e3. |
| [118] |
Deng ZQ, Maksaev G, Schlegel AM, et al. Structural mechanism for gating of a eukaryotic mechanosensitive channel of small conductance[J]. Nat Commun, 2020, 11(1):3690.
doi: 10.1038/s41467-020-17538-1 URL |
| [119] |
Clark MD, Contreras GF, Shen R, et al. Electromechanical coupling in the hyperpolarization-activated K + channel KAT1[J]. Nature, 2020, 583(7814):145-149.
doi: 10.1038/s41586-020-2335-4 URL |
| [120] |
Li SY, Yang F, Sun DM, et al. Cryo-EM structure of the hyperpolarization-activated inwardly rectifying potassium channel KAT1 from Arabidopsis[J]. Cell Res, 2020, 30(11):1049-1052.
doi: 10.1038/s41422-020-00407-3 URL |
| [121] |
Gao F, Han XW, Wu JH, et al. A heat-activated calcium-permeable channel—Arabidopsis cyclic nucleotide-gated ion channel 6—is involved in heat shock responses[J]. Plant J, 2012, 70(6):1056-1069.
doi: 10.1111/tpj.2012.70.issue-6 URL |
| [122] |
Ma Y, Dai XY, Xu YY, et al. COLD1 confers chilling tolerance in rice[J]. Cell, 2015, 160(6):1209-1221.
doi: 10.1016/j.cell.2015.01.046 URL |
| [123] |
Shi YT, Yang SH. COLD1:a cold sensor in rice[J]. Sci China Life Sci, 2015, 58(4):409-410.
doi: 10.1007/s11427-015-4831-6 URL |
| [124] |
Yang TB, Chaudhuri S, Yang LH, et al. A calcium/calmodulin-regulated member of the receptor-like kinase family confers cold tolerance in plants[J]. J Biol Chem, 2010, 285(10):7119-7126.
doi: 10.1074/jbc.M109.035659 URL |
| [125] |
Yang TB, Shad Ali G, Yang LH, et al. Calcium/calmodulin-regulated receptor-like kinase CRLK1 interacts with MEKK1 in plants[J]. Plant Signal Behav, 2010, 5(8):991-994.
doi: 10.4161/psb.5.8.12225 URL |
| [126] | Liu ZY, Jia YX, Ding YL, et al. Plasma membrane CRPK1-mediated phosphorylation of 14-3-3 proteins induces their nuclear import to fine-tune CBF signaling during cold response[J]. Mol Cell, 2017, 66(1): 117-128. e5. |
| [127] | Feng W, Kita D, Peaucelle A, et al. The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling[J]. Curr Biol, 2018, 28(5):666-675. e5. |
| [128] |
Tunyasuvunakool K, Adler J, Wu Z, et al. Highly accurate protein structure prediction for the human proteome[J]. Nature, 2021, 596(7873):590-596.
doi: 10.1038/s41586-021-03828-1 URL |
| [1] | YANG Zhi-xiao, HOU Qian, LIU Guo-quan, LU Zhi-gang, CAO Yi, GOU Jian-yu, WANG Yi, LIN Ying-chao. Responses of Rubisco and Rubisco Activase in Different Resistant Tobacco Strains to Brown Spot Stress [J]. Biotechnology Bulletin, 2023, 39(9): 202-212. |
| [2] | LIU Bao-cai, CHEN Jing-ying, ZHANG Wu-jun, HUANG Ying-zhen, ZHAO Yun-qing, LIU Jian-chao, WEI Zhi-cheng. Characteristics Analysis of Seed Microrhizome Gene Expression of Polygonatum cyrtonema [J]. Biotechnology Bulletin, 2023, 39(8): 220-233. |
| [3] | LIU Kui, LI Xing-fen, YANG Pei-xin, ZHONG Zhao-chen, CAO Yi-bo, ZHANG Ling-yun. Functional Study and Validation of Transcriptional Coactivator PwMBF1c in Picea wilsonii [J]. Biotechnology Bulletin, 2023, 39(5): 205-216. |
| [4] | WEI Ming WANG Xin-yu WU Guo-qiang ZHAO Meng. The Role of NAD-dependent Deacetylase SRT in Plant Epigenetic Inheritance Regulation [J]. Biotechnology Bulletin, 2023, 39(4): 59-70. |
| [5] | WANG Qi, HU Zhe, FU Wei, LI Guang-zhe, HAO Lin. Regulation of Burkholderia sp. GD17 on the Drought Tolerance of Cucumber Seedlings [J]. Biotechnology Bulletin, 2023, 39(3): 163-175. |
| [6] | YAN Xiong-ying, WANG Zhen, WANG Xia, YANG Shi-hui. Microbial Sulfur Metabolism and Stress Resistance [J]. Biotechnology Bulletin, 2023, 39(11): 150-167. |
| [7] | ZHANG Hong-hong, FANG Xiao-feng. Advances in the Regulation of Stress Sensing and Responses by Phase Separation in Plants [J]. Biotechnology Bulletin, 2023, 39(11): 44-53. |
| [8] | YAN Meng-yu, WEI Xiao-wei, CAO Jing, LAN Hai-yan. Cloning of Basic Helix-loop-helix(bHLH)Transcription Factor Gene SabHLH169 in Suaeda aralocaspica and Analysis of Its Resistances to Drought Stress [J]. Biotechnology Bulletin, 2023, 39(11): 328-339. |
| [9] | LIU Yuan-yuan, WEI Chuan-zheng, XIE Yong-bo, TONG Zong-jun, HAN Xing, GAN Bing-cheng, XIE Bao-gui, YAN Jun-jie. Characteristics of Class II Peroxidase Gene Expression During Fruiting Body Development and Stress Response in Flammulina filiformis [J]. Biotechnology Bulletin, 2023, 39(11): 340-349. |
| [10] | RUAN Hang, DUO Hao-yuan, FAN Wen-yan, LV Qing-han, JIANG Shu-jun, ZHU Sheng-wei. Role of the AtERF49 in the Responses to Salt-alkali Stress in Arabidopsis [J]. Biotechnology Bulletin, 2023, 39(1): 150-156. |
| [11] | LI Jian-jian, HE Chen-jing, HUANG Xiao-ping, XIANG Tai-he. Research Progress in the Regulation of Development and Stress Response by Long Non-coding RNAs in Plants [J]. Biotechnology Bulletin, 2023, 39(1): 48-58. |
| [12] | WANG Nan-nan, WANG Wen-jia, ZHU Qiang. Research Progress of microRNAs in Plant Stress Responses [J]. Biotechnology Bulletin, 2022, 38(8): 1-11. |
| [13] | TANG Qian-qian, LIN Chu-yu, TAO Zeng. Research Progress in Histone Demethylase in Plant [J]. Biotechnology Bulletin, 2022, 38(7): 13-22. |
| [14] | GU Pan, QI Xue-ying, LI Li, ZHANG Xi, SHAN Xiao-yi. Endocytosis of AtRGS1 Involved in the Regulation of G-protein-mediated Arabidopsis Development and Stress Responses [J]. Biotechnology Bulletin, 2022, 38(6): 34-42. |
| [15] | ZU Guo-qiang, HU Zhe, WANG Qi, LI Guang-zhe, HAO Lin. Regulatory Role of Burkholderia sp. GD17 in Rice Seedling’s Responses to Cadmium Stress [J]. Biotechnology Bulletin, 2022, 38(4): 153-162. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||