








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (1): 289-298.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0516
Previous Articles Next Articles
ZHANG Xue( ), TAN Yu-meng, JIANG Hai-xia, YANG Guang-yu(
), TAN Yu-meng, JIANG Hai-xia, YANG Guang-yu( )
)
Received:2021-04-15
Online:2022-01-26
Published:2022-02-22
Contact:
YANG Guang-yu
E-mail:yukimura0214@sjtu.edu.cn;yanggy@sjtu.edu.cn
ZHANG Xue, TAN Yu-meng, JIANG Hai-xia, YANG Guang-yu. Directed Evolution of α-1,2-fucosyltransferase by a Single-cell Ultra-high-throughput Screening Method[J]. Biotechnology Bulletin, 2022, 38(1): 289-298.
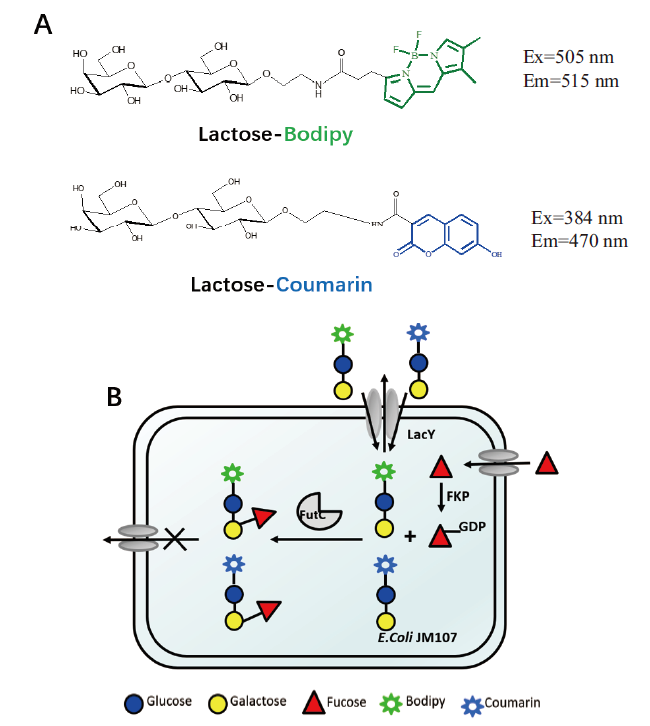
Fig. 1 Fluorescent substrate structure and the schematic diagram of FACS screening strategy A:The fluorescent substrates. B:Schematic diagram of single cell intracellular fluorescence capture
| 引物Primer | 序列Sequence(5'-3') |
|---|---|
| FutC-ep -F | CGAGCTCGACGATGACGATAAAATGGCCTTTAA |
| FutC-ep- R | CCCAAGCTTGGGTCAATCTAAAGCGTTATAC |
Table 1 Primers for random mutation library construction
| 引物Primer | 序列Sequence(5'-3') |
|---|---|
| FutC-ep -F | CGAGCTCGACGATGACGATAAAATGGCCTTTAA |
| FutC-ep- R | CCCAAGCTTGGGTCAATCTAAAGCGTTATAC |
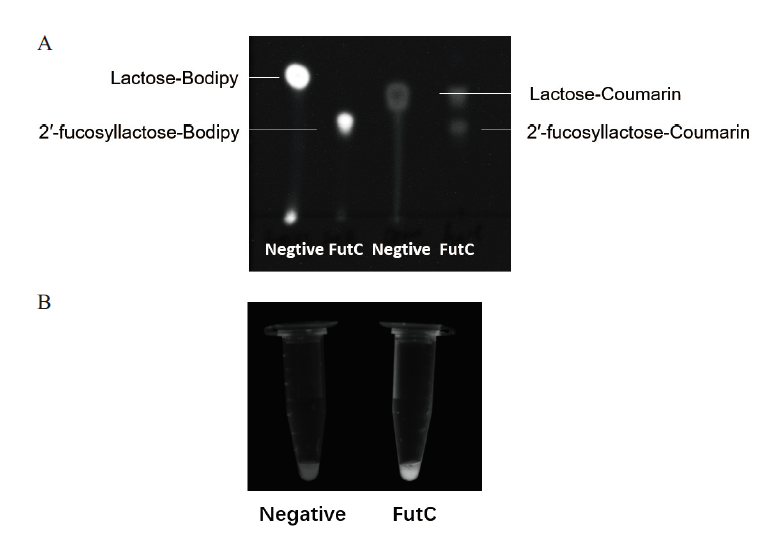
Fig. 2 TLC analysis of fluorescent products and intra-cellular fluorescence enrichment reaction analysis of negative and positive strains under UV light irradiation A:TLC analysis of the two fluorescent substrates(Negtive)and the fluorescent products catalyzed by FutC(FutC). B:Comparison of the intracellular fluorescence enrichment reaction of the negative strain and the positive strain under UV light irradiation,the negative strain(Negtive,JM107 Nan- LacZ- pUCKC18-FKP/pUC18)and positive strains(FutC,JM107 Nan- LacZ- pUCKC18-FKP/pUC18-FutC)
| 分选前阳性比例 Proportion of positive cells before sorting/% | 分选后阳性比例 Proportion of positive cells after sorting/% | 富集倍数 Enrichment factor |
|---|---|---|
| 50 | 100 | 2 |
| 10 | 100 | 10 |
| 1 | 90 | 90 |
| 0.1 | 73.3 | 733 |
Table 2 Model screening of FutC
| 分选前阳性比例 Proportion of positive cells before sorting/% | 分选后阳性比例 Proportion of positive cells after sorting/% | 富集倍数 Enrichment factor |
|---|---|---|
| 50 | 100 | 2 |
| 10 | 100 | 10 |
| 1 | 90 | 90 |
| 0.1 | 73.3 | 733 |
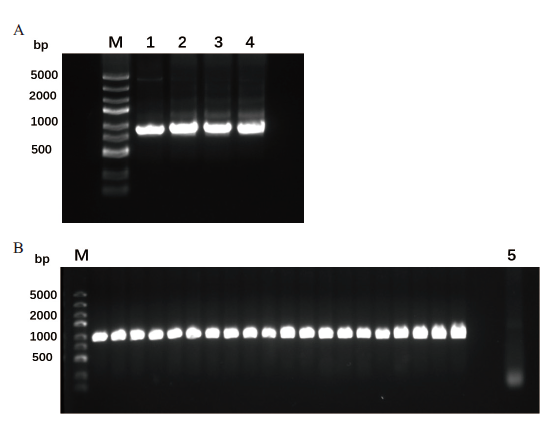
Fig. 5 Construction of the FutC random mutation library A:Error-prone PCR products of futC gene with different Mn2+ concentration,1, 2, 3, 4 are:PCR products with 0, 0.04, 0.06, and 0.1 mmol/L Mn2+. B:Ligation efficiency of random mutation library,and 5 is a negative control
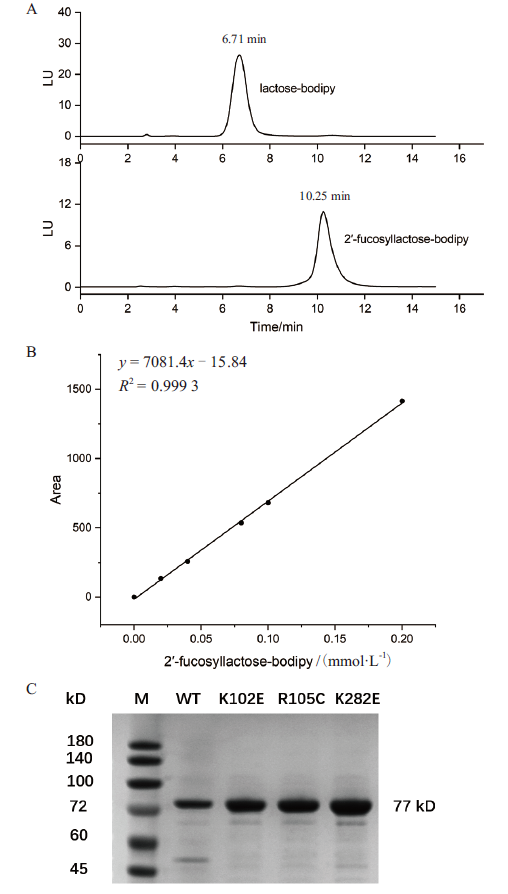
Fig. 8 Establishment of HPLC enzyme activity determin-ation method and protein purification of FutC WT and mutant A:HPLC analysis of substrate lactose-bodipy and product 2'-fucosyllactose-bodipy. B:Standard curve for the quantification of product 2'-fucosyllactose-bodipy. C:SDS-PAGE analysis of purified FutC wild-type and mutant proteins,WT:wild type,1:K102E, 2:R105C,3:K282E
| 酶Enzyme | 突变位点Mutation | 比活力Specific activity/(mU·mg-1) |
|---|---|---|
| WT | None | 57.6±3.6 |
| 1 | K102E | 156.8±2.9 |
| 2 | R105C | 175.7±11.0 |
| 3 | K282E | 151.9±7.9 |
Table 3 Specific activities of WT FutC and selected mutants using lactose-bodipy as acceptor
| 酶Enzyme | 突变位点Mutation | 比活力Specific activity/(mU·mg-1) |
|---|---|---|
| WT | None | 57.6±3.6 |
| 1 | K102E | 156.8±2.9 |
| 2 | R105C | 175.7±11.0 |
| 3 | K282E | 151.9±7.9 |
| [1] |
Tu Z, Lin YN, Lin CH. Development of fucosyltransferase and fucosidase inhibitors[J]. Chem Soc Rev, 2013, 42(10):4459-4475.
doi: 10.1039/c3cs60056d URL |
| [2] |
Keeley TS, Yang SY, Lau E. The diverse contributions of fucose linkages in cancer[J]. Cancers, 2019, 11(9):1241.
doi: 10.3390/cancers11091241 URL |
| [3] |
Thomas D, Rathinavel AK, Radhakrishnan P. Altered glycosylation in cancer:a promising target for biomarkers and therapeutics[J]. Biochim Biophys Acta Rev Cancer, 2021, 1875(1):188464.
doi: 10.1016/j.bbcan.2020.188464 URL |
| [4] |
Danishefsky SJ, Shue YK, Chang MN, et al. Development of Globo-H cancer vaccine[J]. Acc Chem Res, 2015, 48(3):643-652.
doi: 10.1021/ar5004187 URL |
| [5] |
Huang CS, Yu AL, Tseng LM, et al. Globo H-KLH vaccine adagloxad simolenin(OBI-822)/OBI-821 in patients with metastatic breast cancer:phase II randomized, placebo-controlled study[J]. J Immunother Cancer, 2020, 8(2):e000342.
doi: 10.1136/jitc-2019-000342 URL |
| [6] |
Guo J, Jiang W, Li Q, et al. Comparative immunological studies of tumor-associated Lewis X, Lewis Y, and KH-1 antigens[J]. Carbohydr Res, 2020, 492:107999.
doi: 10.1016/j.carres.2020.107999 URL |
| [7] |
Bode L. Human milk oligosaccharides:every baby needs a sugar Mama[J]. Glycobiology, 2012, 22(9):1147-1162.
doi: 10.1093/glycob/cws074 URL |
| [8] |
He Y, Liu S, Kling DE, et al. The human milk oligosaccharide 2'-fucosyllactose modulates CD14 expression in human enterocytes, thereby attenuating LPS-induced inflammation[J]. Gut, 2016, 65(1):33-46.
doi: 10.1136/gutjnl-2014-307544 URL |
| [9] |
Orczyk-Pawiłowicz M, Lis-Kuberka J. The impact of dietary fucosylated oligosaccharides and glycoproteins of human milk on infant well-being[J]. Nutrients, 2020, 12(4):1105.
doi: 10.3390/nu12041105 URL |
| [10] | Sprenger N, Binia A, Austin S. Human milk oligosaccharides:factors affecting their composition and their physiological significance[J]. Nestle Nutr Inst Work Ser, 2019, 90:43-56. |
| [11] |
Goehring KC, Marriage BJ, Oliver JS, et al. Similar to those who are breastfed, infants fed a formula containing 2'-fucosyllactose have lower inflammatory cytokines in a randomized controlled trial[J]. J Nutr, 2016, 146(12):2559-2566.
pmid: 27798337 |
| [12] |
Musilova S, Rada V, Vlkova E, et al. Beneficial effects of human milk oligosaccharides on gut microbiota[J]. Benef Microbes, 2014, 5(3):273-283.
doi: 10.3920/BM2013.0080 pmid: 24913838 |
| [13] | Zhu Y, Wan L, Li W, et al. Recent advances on 2'-fucosyllactose:physiological properties, applications, and production approaches[J]. Crit Rev Food Sci Nutr, 2020:1-10. |
| [14] |
Zhou WT, Jiang H, Wang LL, et al. Biotechnological production of 2'-fucosyllactose:a prevalent fucosylated human milk oligosaccharide[J]. ACS Synth Biol, 2021, 10(3):447-458.
doi: 10.1021/acssynbio.0c00645 URL |
| [15] |
Drouillard S, Driguez H, Samain E. Large-scale synjournal of H-antigen oligosaccharides by expressing Helicobacter pylori alpha1, 2-fucosyltransferase in metabolically engineered Escherichia coli cells[J]. Angew Chem Int Ed Engl, 2006, 45(11):1778-1780.
doi: 10.1002/(ISSN)1521-3773 URL |
| [16] |
Chin YW, Kim JY, Lee WH, et al. Enhanced production of 2'-fucosyllactose in engineered Escherichia coli BL21star(DE3)by modulation of lactose metabolism and fucosyltransferase[J]. J Biotechnol, 2015, 210:107-115.
doi: 10.1016/j.jbiotec.2015.06.431 URL |
| [17] |
Huang D, Yang K, Liu J, et al. Metabolic engineering of Escherichia coli for the production of 2'-fucosyllactose and 3-fucosyllactose through modular pathway enhancement[J]. Metab Eng, 2017, 41:23-38.
doi: S1096-7176(16)30233-6 pmid: 28286292 |
| [18] |
Arnold FH, Volkov AA. Directed evolution of biocatalysts[J]. Curr Opin Chem Biol, 1999, 3(1):54-59.
pmid: 10021399 |
| [19] |
Turner NJ. Directed evolution drives the next generation of biocatalysts[J]. Nat Chem Biol, 2009, 5(8):567-573.
doi: 10.1038/nchembio.203 URL |
| [20] |
Aharoni A, Thieme K, Chiu CP, et al. High-throughput screening methodology for the directed evolution of glycosyltransferases[J]. Nat Methods, 2006, 3(8):609-614.
pmid: 16862135 |
| [21] |
Yang GY, Rich JR, Gilbert M, et al. Fluorescence activated cell sorting as a general ultra-high-throughput screening method for directed evolution of glycosyltransferases[J]. J Am Chem Soc, 2010, 132(30):10570-10577.
doi: 10.1021/ja104167y URL |
| [22] |
Tan YM, Zhang Y, Han YB, et al. Directed evolution of an α 1, 3-fucosyltransferase using a single-cell ultrahigh-throughput screening method[J]. Sci Adv, 2019, 5(10):eaaw8451. DOI: 10.1126/sciadv.aaw8451.
doi: 10.1126/sciadv.aaw8451 |
| [23] |
Stein D, Lin YN, Lin CH. Characterization of Helicobacter pylori α1, 2-fucosyltransferase for enzymatic synjournal of tumor-associated antigens[J]. Adv Synth Catal, 2008, 350(14/15):2313-2321.
doi: 10.1002/adsc.v350:14/15 URL |
| [24] |
Engels L, Elling L. WbgL:a novel bacterial α1, 2-fucosyltransferase for the synjournal of 2'-fucosyllactose[J]. Glycobiology, 2014, 24(2):170-178.
doi: 10.1093/glycob/cwt096 URL |
| [25] |
Li C, Wu M, Gao X, et al. Efficient biosynjournal of 2'-fucosyllactose using an in vitro multienzyme cascade[J]. J Agric Food Chem, 2020, 68(39):10763-10771.
doi: 10.1021/acs.jafc.0c04221 URL |
| [1] | QU Ge, SUN Zhou-tong. Catalytic Promiscuity-driven Redesign of Enzyme Functions [J]. Biotechnology Bulletin, 2023, 39(4): 1-9. |
| [2] | WANG Mu-qiang, CHEN Qi, MA Wei, LI Chun-xiu, OUYANG Peng-fei, XU Jian-he. Advances in the Application of Machine Learning Methods for Directed Evolution of Enzymes [J]. Biotechnology Bulletin, 2023, 39(4): 38-48. |
| [3] | ZHANG Chen, ZHANG Tong-tong, LIU Hai-ping. Screening and Identification of Ethylene-forming Enzymes with High Activity and Thermostability [J]. Biotechnology Bulletin, 2022, 38(11): 269-276. |
| [4] | CHEN Chun, SU Ling-qia, XIA Wei, WU Jing. Improved the Thermostability of MTHase from Arthrobacter ramosus by Directed Evolution [J]. Biotechnology Bulletin, 2021, 37(3): 84-91. |
| [5] | REN Tian-lei, YANG Hai-quan, XU Fei. Directed Evolution of Methyl Parathion Hydrolase Based on the Multi-dimensional Features:Molecular Structure and Bioinformatics [J]. Biotechnology Bulletin, 2018, 34(10): 194-200. |
| [6] | WANG Xiao-lu, WANG Yu, LIU Jiao,ZHENG Ping,LU Fu-ping. Enhanced Methanol Utilization in Genetically Engineered Escherichia coli by Directed Evolution [J]. Biotechnology Bulletin, 2017, 33(9): 101-109. |
| [7] | GUO Yuan, ZHAO Zhong-lin. Advances on Applications of Synthetic Biology and Directed Evolution in Microbial Systems [J]. Biotechnology Bulletin, 2017, 33(1): 76-82. |
| [8] | ZHANG Xue-ling CHEN Xiao-li LI He. Determination of Enzymatic Properties of a Laccase Lac1338,and Effects of Directed Mutants on the Degradations of Different Dyes [J]. Biotechnology Bulletin, 2016, 32(7): 170-177. |
| [9] | Lü Yongkun, Du Guocheng, Chen Jian, Zhou Jingwen. Advances in Synthetic Biology [J]. Biotechnology Bulletin, 2015, 31(4): 134-148. |
| [10] | Liu Miao, Wang Yonghui, Lu Qun. Advances in Directed Research of VD3 Hydroxylase [J]. Biotechnology Bulletin, 2014, 0(1): 27-31. |
| [11] | Shao Min, Li Changfu, Ge Zhenglong, Zhou Hefeng. Directed evolution of β-glucanase from Bacillus subtilis by Error-prone PCR [J]. Biotechnology Bulletin, 2013, 0(12): 141-145. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||