








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (6): 272-278.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1432
Previous Articles Next Articles
LIU Jing-jing( ), LIU Xiao-rui, LI Lin, WANG Ying, YANG Hai-yuan(
), LIU Xiao-rui, LI Lin, WANG Ying, YANG Hai-yuan( ), DAI Yi-fan(
), DAI Yi-fan( )
)
Received:2021-11-15
Online:2022-06-26
Published:2022-07-11
Contact:
YANG Hai-yuan,DAI Yi-fan
E-mail:564665840@qq.com;hyyang@njmu.edu.cn;daiyifan@njmu.edu.cn
LIU Jing-jing, LIU Xiao-rui, LI Lin, WANG Ying, YANG Hai-yuan, DAI Yi-fan. Establishment of Porcine Fetal Fibroblasts with OXTR-knockout Using CRISPR/Cas9[J]. Biotechnology Bulletin, 2022, 38(6): 272-278.
| 名称 Primer name | 引物序列 Primer sequence(5'-3') | 产物大小 Fragment length |
|---|---|---|
| OXTR-F | ACCTGCCCAAGAAGTCTCAG | 748 bp |
| OXTR-R | CAGCAGCAGGTAAGTGGAAG |
Table 1 Amplification primers for exon 2 sequene of OXTR gene
| 名称 Primer name | 引物序列 Primer sequence(5'-3') | 产物大小 Fragment length |
|---|---|---|
| OXTR-F | ACCTGCCCAAGAAGTCTCAG | 748 bp |
| OXTR-R | CAGCAGCAGGTAAGTGGAAG |

Fig.2 Analysis of primary,secondary,and tertiary structures of OXTR between humans and pigs A:Amino acid sequence homology analysis of human and pig OXTR,and the identical amino acids are blue shaded. B:Secondary structures of human and pig proteins. Red indicates alpha-helix;green indicates beta-sheet;and blue indicates beta-turn. C:Tertiary structures of human and pig proteins. Red indicates the structure of human OXTR,and green indicates the structure of pig OXTR
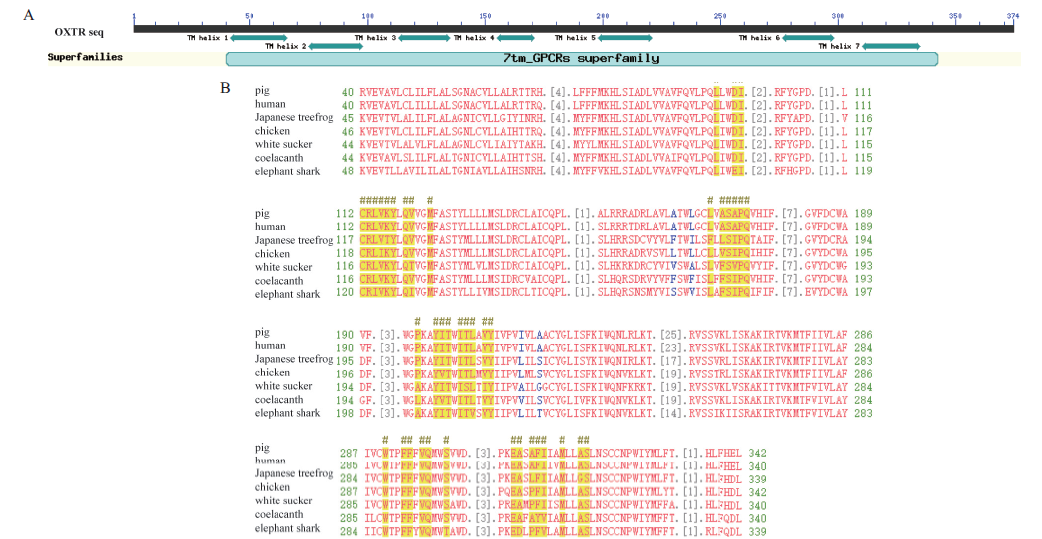
Fig. 3 Identification of domains and key residues of porcine OXTR A:Schematic diagram of domains and catalytic residues of pig OXTR. B:Multiple sequence alignment of OXTR,# denotes critical amino acid residues binding to ligand
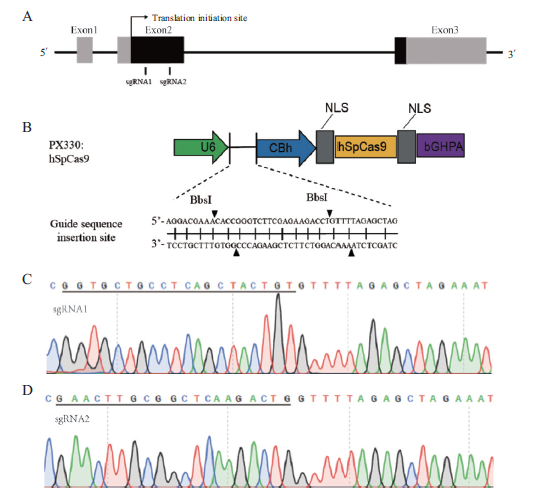
Fig.4 Target of OXTR gene and sequencing of recombinant vectors A:Schematic diagram of OXTR gene target in Bama miniature pig. Gray indicates the non-coding area and black indicates the coding areas. B:Schematic diagram of pX330 vector. C and D:Sequencing map of recombinant vector. Black underlined is the insertion sequence
| sgRNA | 序列Sequence(5'-3') |
|---|---|
| sgRNA-1-F | CACCGGTGCTGCCTCAGCTACTGT |
| sgRNA-1-R | AAACACAGTAGCTGAGGCAGCACC |
| sgRNA-2-F | CACCGAACTTGCGGCTCAAGACTG |
| sgRNA-2-R | AAACCAGTCTTGAGCCGCAAGTTC |
Table 2 Oligonucleotide sequences of sgRNAs at OXTR targeting sites
| sgRNA | 序列Sequence(5'-3') |
|---|---|
| sgRNA-1-F | CACCGGTGCTGCCTCAGCTACTGT |
| sgRNA-1-R | AAACACAGTAGCTGAGGCAGCACC |
| sgRNA-2-F | CACCGAACTTGCGGCTCAAGACTG |
| sgRNA-2-R | AAACCAGTCTTGAGCCGCAAGTTC |
| 编号 ID name | 目的片段 Target fragment | 突变型 Indels |
|---|---|---|
| WT | CAGGTGCTGCCTCAGCTACTGTGGG//GGCAGAACTTGCGGCTCAAGACTGCG | WT |
| 1 | CAGGTGCTGCCTCAGCTAC-----// ----------------------CTGCGG | -411 bp |
| 2 | CAGGTGCTGCCTCAGCTAC-----// ----------------------CTGCGG | -411 bp |
| 3 | CAGGTGCTGCCTCAGCTAC-----// ----------------------CTGCGG | -411 bp |
| 4 | CAGGTGCTGCCTCAGCTACTG---// -------------------------CGG | -412 bp |
| 5 | CAGGTGCTGCCTCAGCTACTCTTGA//AGCGGAAGGTGATATCCCACACTGCGG | -289,+289 bp |
Table 3 Genotypes of OXTR-knockout PFFs
| 编号 ID name | 目的片段 Target fragment | 突变型 Indels |
|---|---|---|
| WT | CAGGTGCTGCCTCAGCTACTGTGGG//GGCAGAACTTGCGGCTCAAGACTGCG | WT |
| 1 | CAGGTGCTGCCTCAGCTAC-----// ----------------------CTGCGG | -411 bp |
| 2 | CAGGTGCTGCCTCAGCTAC-----// ----------------------CTGCGG | -411 bp |
| 3 | CAGGTGCTGCCTCAGCTAC-----// ----------------------CTGCGG | -411 bp |
| 4 | CAGGTGCTGCCTCAGCTACTG---// -------------------------CGG | -412 bp |
| 5 | CAGGTGCTGCCTCAGCTACTCTTGA//AGCGGAAGGTGATATCCCACACTGCGG | -289,+289 bp |
| [1] |
Park HR, Lee JM, Moon HE, et al. A short review on the current understanding of autism spectrum disorders[J]. Exp Neurobiol, 2016, 25(1):1-13.
doi: 10.5607/en.2016.25.1.1 pmid: 26924928 |
| [2] |
Feldman R, Monakhov M, Pratt M, et al. Oxytocin pathway genes:evolutionary ancient system impacting on human affiliation, sociality, and psychopathology[J]. Biol Psychiatry, 2016, 79(3):174-184.
doi: 10.1016/j.biopsych.2015.08.008 URL |
| [3] |
Tops S, Habel U, Radke S. Genetic and epigenetic regulatory mechanisms of the oxytocin receptor gene(OXTR)and the(clinical)implications for social behavior[J]. Horm Behav, 2019, 108:84-93.
doi: 10.1016/j.yhbeh.2018.03.002 URL |
| [4] |
Jurek B, Neumann ID. The oxytocin receptor:from intracellular signaling to behavior[J]. Physiol Rev, 2018, 98(3):1805-1908.
doi: 10.1152/physrev.00031.2017 URL |
| [5] | Vaidyanathan R, Hammock EAD. Oxytocin receptor gene loss influences expression of the oxytocin gene in C57BL/6J mice in a sex- and age-dependent manner[J]. J Neuroendocrinol, 2020, 32(2):e12821. |
| [6] |
Haram M, Bettella F, Brandt CL, et al. Contribution of oxytocin receptor polymorphisms to amygdala activation in schizophrenia spectrum disorders[J]. BJPsych Open, 2016, 2(6):353-358.
doi: 10.1192/bjpo.bp.116.003376 URL |
| [7] |
Uzefovsky F, Bethlehem RAI, Shamay-Tsoory S, et al. The oxytocin receptor gene predicts brain activity during an emotion recognition task in autism[J]. Mol Autism, 2019, 10:12.
doi: 10.1186/s13229-019-0258-4 pmid: 30918622 |
| [8] |
Caria A, Ciringione L, Falco S. Morphofunctional alterations of the hypothalamus and social behavior in autism spectrum disorders[J]. Brain Sci, 2020, 10(7):435.
doi: 10.3390/brainsci10070435 URL |
| [9] |
Pobbe RLH, Pearson BL, Defensor EB, et al. Oxytocin receptor knockout mice display deficits in the expression of autism-related behaviors[J]. Horm Behav, 2012, 61(3):436-444.
doi: 10.1016/j.yhbeh.2011.10.010 URL |
| [10] | Ribeiro D, Nunes AR, Gliksberg M, et al. Oxytocin receptor signalling modulates novelty recognition but not social preference in zebrafish[J]. J Neuroendocrinol, 2020, 32(4):e12834. |
| [11] |
Chang SWC, Brent LJN, Adams GK, et al. Neuroethology of primate social behavior[J]. Proc Natl Acad Sci USA, 2013, 110(Suppl 2):10387-10394.
doi: 10.1073/pnas.1301213110 URL |
| [12] | Ma YL, Wei JL, Zhang Q, et al. A genome scan for selection signatures in pigs[J]. PLoS One, 2015, 10(3):e0116850. |
| [13] |
Li MZ, Chen L, Tian SL, et al. Comprehensive variation discovery and recovery of missing sequence in the pig genome using multiple de novo assemblies[J]. Genome Res, 2017, 27(5):865-874.
doi: 10.1101/gr.207456.116 URL |
| [14] |
Zhang JF, Khazalwa EM, Abkallo HM, et al. The advancements, challenges, and future implications of the CRISPR/Cas9 system in swine research[J]. J Genet Genomics, 2021, 48(5):347-360.
doi: 10.1016/j.jgg.2021.03.015 URL |
| [15] |
Sauleau P, Lapouble E, Val-Laillet D, et al. The pig model in brain imaging and neurosurgery[J]. Animal, 2009, 3(8):1138-1151.
doi: 10.1017/S1751731109004649 pmid: 22444844 |
| [16] |
Gao MY, Zhu XL, Yang G, et al. CRISPR/Cas9-mediated gene editing in porcine models for medical research[J]. DNA Cell Biol, 2021, 40(12):1462-1475.
doi: 10.1089/dna.2020.6474 URL |
| [17] |
Yang W, Chen X, Li S, et al. Genetically modified large animal models for investigating neurodegenerative diseases[J]. Cell Biosci, 2021, 11(1):218.
doi: 10.1186/s13578-021-00729-8 URL |
| [18] |
Gimpl G, Fahrenholz F. The oxytocin receptor system:structure, function, and regulation[J]. Physiol Rev, 2001, 81(2):629-683.
pmid: 11274341 |
| [19] |
Tick B, Bolton P, Happé F, et al. Heritability of autism spectrum disorders:a meta-analysis of twin studies[J]. J Child Psychol Psychiatry, 2016, 57(5):585-595.
doi: 10.1111/jcpp.12499 URL |
| [20] |
Liu J, Nyholt DR, Magnussen P, et al. A genomewide screen for autism susceptibility loci[J]. Am J Hum Genet, 2001, 69(2):327-340.
doi: 10.1086/321980 pmid: 11452361 |
| [21] |
Meyer-Lindenberg A, Tost H. Neural mechanisms of social risk for psychiatric disorders[J]. Nat Neurosci, 2012, 15(5):663-668.
doi: 10.1038/nn.3083 pmid: 22504349 |
| [22] |
Cataldo I, Azhari A, Esposito G. A review of oxytocin and arginine-vasopressin receptors and their modulation of autism spectrum disorder[J]. Front Mol Neurosci, 2018, 11:27.
doi: 10.3389/fnmol.2018.00027 pmid: 29487501 |
| [23] |
Li Z, Yang HY, Wang Y, et al. Generation of tryptophan hydroxylase 2 gene knockout pigs by CRISPR/Cas9-mediated gene targeting[J]. J Biomed Res, 2017, 31(5):445-452.
doi: 10.7555/JBR.31.20170026 URL |
| [24] |
Yao J, Zeng HS, Zhang M, et al. OSBPL2-disrupted pigs recapitulate dual features of human hearing loss and hypercholesterolaemia[J]. J Genet Genomics, 2019, 46(8):379-387.
doi: 10.1016/j.jgg.2019.06.006 URL |
| [25] |
Denes CE, Cole AJ, Aksoy YA, et al. Approaches to enhance precise CRISPR/Cas9-mediated genome editing[J]. Int J Mol Sci, 2021, 22(16):8571.
doi: 10.3390/ijms22168571 URL |
| [1] | CHEN Xiao-ling, LIAO Dong-qing, HUANG Shang-fei, CHEN Ying, LU Zhi-long, CHEN Dong. Advances in CRISPR/Cas9 System Modifying Saccharomycescerevisiae [J]. Biotechnology Bulletin, 2023, 39(8): 148-158. |
| [2] | YANG Yu-mei, ZHANG Kun-xiao. Establishing a Stable Cell Line with Site-specific Integration of ERK Kinase Phase-separated Fluorescent Probe Using CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(8): 159-164. |
| [3] | SHI Wei-tao, YAO Chun-peng, WEI Wen-Kang, WANG Lei, FANG Yuan-jie, TONG Yu-jie, MA Xiao-jiao, JIANG Wen, ZHANG Xiao-ai, SHAO Wei. Establishment of MDH2 Knockout Cell Line Using CRISPR/Cas9 Technology and Study of Anti-deoxynivalenol Effect [J]. Biotechnology Bulletin, 2023, 39(7): 307-315. |
| [4] | LIU Xiao-yan, ZHU Zhen-liang, SHI Guang-yu, HUA Zi-yu, YANG Chen, ZHANG Yong, LIU Jun. Strategies to Optimize the Expression of Mammary Gland Bioreactor [J]. Biotechnology Bulletin, 2023, 39(5): 77-91. |
| [5] | WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(4): 246-258. |
| [6] | PING Huai-lei, GUO Xue, YU Xiao, SONG Jing, DU Chun, WANG Juan, ZHANG Huai-bi. Cloning and Expression of PdANS in Paeonia delavayi and Correlation with Anthocyanin Content [J]. Biotechnology Bulletin, 2023, 39(3): 206-217. |
| [7] | CHENG Jing-wen, CAO Lei, ZHANG Yan-min, YE Qian, CHEN Min, TAN Wen-song, ZHAO Liang. Establishment and Application of Multigene Engineering Transformation Strategy for CHO Cells [J]. Biotechnology Bulletin, 2023, 39(2): 283-291. |
| [8] | HUANG Wen-li, LI Xiang-xiang, ZHOU Wen-ting, LUO Sha, YAO Wei-jia, MA Jie, ZHANG Fen, SHEN Yu-sen, GU Hong-hui, WANG Jian-sheng, SUN Bo. Targeted Editing of BoZDS in Broccoli by CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(2): 80-87. |
| [9] | WANG Bing, ZHAO Hui-na, YU Jing, CHEN Jie, LUO Mei, LEI Bo. Regulation of Leaf Bud by REVOLUTA in Tobacco Based on CRISPR/Cas9 System [J]. Biotechnology Bulletin, 2023, 39(10): 197-208. |
| [10] | LI Shuang-xi, HUA Jin-lian. Research Progress in Anti-porcine Reproductive and Respiratory Syndrome Genetically Modified Pigs [J]. Biotechnology Bulletin, 2023, 39(10): 50-57. |
| [11] | LIN Rong, ZHENG Yue-ping, XU Xue-zhen, LI Dan-dan, ZHENG Zhi-fu. Functional Analysis of ACOL8 Gene in the Ethylene Synthesis and Response in Arabidopsis thaliana [J]. Biotechnology Bulletin, 2023, 39(1): 157-165. |
| [12] | GUO Zhi-hao, JIN Ze-xin, LIU Qi, GAO Li. Bioinformatics Analysis, Subcellular Localization and Toxicity Verification of Effector g11335 in Tilletia contraversa Kühn [J]. Biotechnology Bulletin, 2022, 38(8): 110-117. |
| [13] | YU Qiu-lin, MA Jing-yi, ZHAO Pan, SUN Peng-fang, HE Yu-mei, LIU Shi-biao, GUO Hui-hong. Cloning and Functional Analysis of Gynostemma pentaphyllum GpMIR156a and GpMIR166b [J]. Biotechnology Bulletin, 2022, 38(7): 186-193. |
| [14] | CHEN Jia-min, LIU Yong-jie, MA Jin-xiu, LI Dan, GONG Jie, ZHAO Chang-ping, GENG Hong-wei, GAO Shi-qing. Expression Pattern Analysis of Histone Methyltransferase Under Drought Stress in Hybrid Wheat [J]. Biotechnology Bulletin, 2022, 38(7): 51-61. |
| [15] | WANG Nan, ZHANG Rui, PAN Yang-yang, HE Hong-hong, WANG Jing-lei, CUI Yan, YU Si-jiu. Cloning of Bos grunniens TGF-β1 Gene and Its Expression in Major Organs of Female Reproductive System [J]. Biotechnology Bulletin, 2022, 38(6): 279-290. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||