








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (9): 167-179.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1459
Previous Articles Next Articles
YUAN Xing( ), GUO Cai-hua, LIU Jin-ming, KANG Chao, QUAN Shao-wen, NIU Jian-xin(
), GUO Cai-hua, LIU Jin-ming, KANG Chao, QUAN Shao-wen, NIU Jian-xin( )
)
Received:2021-11-12
Online:2022-09-26
Published:2022-10-11
Contact:
NIU Jian-xin
E-mail:354506832@qq.com;njx105@163.com
YUAN Xing, GUO Cai-hua, LIU Jin-ming, KANG Chao, QUAN Shao-wen, NIU Jian-xin. Genome-wide Identification of CONSTANS-Like Family Genes and Expression Analysis in Wanlut[J]. Biotechnology Bulletin, 2022, 38(9): 167-179.
| 基因 Gene | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|
| JrCOL1 | TTCGAGAAGACGATCCGATACG | CGATCAACTTCCGATCCCATCT |
| JrCOL7 | ATTCCCCTTCTGTGGGTGAATC | TGGCTCATGTTGTTTTCTGCAG |
| JrCOL8 | GTTATGATGCCATCGACAGCAC | TGCCATTCCATTAACGTGAGGA |
| JrCOL23 | CGTGACGGATGTATCGGATTCA | TTTTCTCTGTACCTCAGCACCC |
| JrCOL31 | CTGCAGAAAACAACATGAGCCA | AATGTGAGGCCCAGAATGAACT |
| JrCOL33 | GGTTCACTGGATGTCCTTCTGT | TGAGAAGCATCATCCCAAACCA |
| JrCOL35 | ACCCCAATTTCAGTTCCAAGGT | TATCCGTTCCCGCACTGTAAAA |
| JrCOL42 | GTGGCTCTAAAGATGTCCCACA | TATTCTGTGCCAGTTGCTCCAT |
| JrCOL50 | CTCGTTGCGATCATCTTGTGTC | GATGACACTGAAGAGCCTCCAA |
| JrCOL52 | AGAATTGGGAAATGGCAGGGAT | CCATCGTAAGCAAAGCCGTAAG |
| 18S | GGTCAATCTTCTCGTTCCCTT | TCGCATTTCGCTACGTTCTT |
Table 1 Primer sequences
| 基因 Gene | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|
| JrCOL1 | TTCGAGAAGACGATCCGATACG | CGATCAACTTCCGATCCCATCT |
| JrCOL7 | ATTCCCCTTCTGTGGGTGAATC | TGGCTCATGTTGTTTTCTGCAG |
| JrCOL8 | GTTATGATGCCATCGACAGCAC | TGCCATTCCATTAACGTGAGGA |
| JrCOL23 | CGTGACGGATGTATCGGATTCA | TTTTCTCTGTACCTCAGCACCC |
| JrCOL31 | CTGCAGAAAACAACATGAGCCA | AATGTGAGGCCCAGAATGAACT |
| JrCOL33 | GGTTCACTGGATGTCCTTCTGT | TGAGAAGCATCATCCCAAACCA |
| JrCOL35 | ACCCCAATTTCAGTTCCAAGGT | TATCCGTTCCCGCACTGTAAAA |
| JrCOL42 | GTGGCTCTAAAGATGTCCCACA | TATTCTGTGCCAGTTGCTCCAT |
| JrCOL50 | CTCGTTGCGATCATCTTGTGTC | GATGACACTGAAGAGCCTCCAA |
| JrCOL52 | AGAATTGGGAAATGGCAGGGAT | CCATCGTAAGCAAAGCCGTAAG |
| 18S | GGTCAATCTTCTCGTTCCCTT | TCGCATTTCGCTACGTTCTT |
| 基因名称 Gene name | 基因ID Gene ID | 染色体位置 Chromosome position | 相关蛋白 Related protein | 蛋白重命名 Protein rename | 氨基酸长度 aa length | 蛋白分子量 Molecular weight/kD | 理论等电点pI | 亚细胞定位Subc- ellular localization |
|---|---|---|---|---|---|---|---|---|
| JrCOL1 | LOC109001350 | Chr.1:1256032..1258201 | XP_018834143.1 | JrCOL1 | 373 | 41.31 | 5.17 | 叶绿体 |
| JrCOL2 | LOC109001381 | Chr.1:1488506..1490928 | XP_035544637.1 | JrCOL2 | 211 | 23.40 | 5.73 | 细胞质 |
| JrCOL3 | LOC109009665 | Chr.1:3978995..3983057 | XP_018845790.2 | JrCOL3 | 350 | 39.64 | 8.72 | 细胞核 |
| JrCOL4a-d | LOC109010156 | Chr.1:18366251..18370500 | XP_035543378.1 | JrCOL4a | 428 | 47.24 | 5.27 | 细胞核 |
| XP_018846422.1 | JrCOL4b | 428 | 47.24 | 5.27 | 细胞核 | |||
| XP_018846423.1 | JrCOL4c | 428 | 47.24 | 5.27 | 细胞核 | |||
| XP_018846424.1 | JrCOL4d | 428 | 47.24 | 5.27 | 细胞核 | |||
| JrCOL5 | LOC109015440 | Chr.1:18372462..18372860 | XP_018853453.2 | JrCOL5 | 132 | 14.50 | 6.85 | 细胞核 |
| JrCOL6a-b | LOC108991987 | Chr.1:19317524..19318826 | XP_018821963.2 | JrCOL6a | 237 | 25.91 | 4.42 | 细胞核 |
| XP_018821964.2 | JrCOL6b | 236 | 25.78 | 4.42 | 细胞核 | |||
| JrCOL7a-c | LOC109000761 | Chr.1:44957519..44963229 | XP_018833298.1 | JrCOL7a | 416 | 45.11 | 5.19 | 细胞核 |
| XP_018833299.1 | JrCOL7b | 416 | 45.11 | 5.19 | 细胞核 | |||
| XP_018833303.1 | JrCOL7c | 416 | 45.11 | 5.19 | 细胞核 | |||
| JrCOL8 | LOC109013302 | Chr.2:7141021..7147381 | XP_018850883.1 | JrCOL8 | 539 | 60.48 | 6.22 | 细胞核 |
| JrCOL9 | LOC108984132 | Chr.2:11707792..11709460 | XP_018811512.1 | JrCOL9 | 277 | 31.23 | 4.94 | 细胞核 |
| JrCO10 | LOC109014521 | Chr.2:30680768..30682741 | XP_018852564.1 | JrCOL10 | 333 | 37.49 | 4.97 | 细胞核 |
| JrCOL11a-b | LOC109014085 | Chr.2:35047335..35048788 | XP_018851954.1 | JrCOL11a | 258 | 27.76 | 4.28 | 细胞核 |
| XP_018851962.1 | JrCOL11b | 255 | 27.37 | 4.25 | 细胞核 | |||
| JrCOL12a-b | LOC108999038 | Chr.3:18899526..18901860 | XP_018831379.1 | JrCOL12a | 150 | 16.21 | 4.00 | 细胞核,叶绿体 |
| XP_035544064.1 | JrCOL12b | 150 | 16.21 | 4.00 | 细胞核,叶绿体 | |||
| JrCOL13 | LOC109017893 | Chr.4:14653613..14655876 | XP_035544914.1 | JrCOL13 | 170 | 18.24 | 4.11 | 细胞核 |
| JrCOL14 | LOC108989855 | Chr.5:6297905..6299253 | XP_018819161.1 | JrCOL14 | 220 | 26.37 | 4.94 | 细胞核 |
| JrCOL15a-b | LOC109022307 | Chr.5:9188186..9197431 | XP_018860727.1 | JrCOL15a | 789 | 86.08 | 6.30 | 叶绿体 |
| XP_018860748.1 | JrCOL15b | 778 | 84.89 | 6.35 | 叶绿体 | |||
| JrCOL16 | LOC108994549 | Chr.6:15543376..15545802 | XP_018825341.2 | JrCOL16 | 444 | 48.81 | 5.88 | 细胞核 |
| JrCOL17 | LOC108998057 | Chr.6:29988891..29991120 | XP_018830038.2 | JrCOL17 | 407 | 46.32 | 5.46 | 细胞核 |
| JrCOL18 | LOC108992642 | Chr.7:12544651..12554366 | XP_018822785.2 | JrCOL18 | 685 | 75.72 | 6.71 | 细胞核 |
| JrCOL19 | LOC108983659 | Chr.7:14608063..14609540 | XP_018810913.1 | JrCOL19 | 266 | 28.49 | 8.73 | 叶绿体 |
| JrCOL20 | LOC109005469 | Chr.7:32506953..32517320 | XP_018839979.2 | JrCOL20 | 238 | 26.36 | 5.08 | 细胞核 |
| JrCOL21 | LOC109019524 | Chr.7:38617705..38622031 | XP_018857358.1 | JrCOL21 | 432 | 46.81 | 4.35 | 叶绿体 |
| JrCOL22a-b | LOC109014549 | Chr.7:49644025..49658252 | XP_035548321.1 | JrCOL22a | 780 | 85.75 | 6.42 | 叶绿体 |
| XP_035548322.1 | JrCOL22b | 778 | 85.50 | 6.42 | 叶绿体 | |||
| JrCOL23 | LOC109000996 | Chr.8:600112..601757 | XP_018833641.1 | JrCOL23 | 345 | 37.98 | 6.16 | 细胞质 |
| JrCOL24 | LOC108984911 | Chr.8:6608517..6610109 | XP_018812557.1 | JrCOL24 | 301 | 33.04 | 6.25 | 细胞核 |
| JrCOL25a-b | LOC108980667 | Chr.8:24595513..24597399 | XP_035549568.1 | JrCOL25a | 158 | 17.12 | 5.62 | 细胞质 |
| XP_035549569.1 | JrCOL25b | 117 | 12.99 | 6.49 | 细胞质 | |||
| JrCOL26 | LOC108997344 | Chr.9:19840907..19842615 | XP_018829103.1 | JrCOL26 | 331 | 37.53 | 5.83 | 细胞核 |
| JrCOL27 | LOC108985349 | Chr.9:21400481..21403221 | XP_018813169.1 | JrCOL27 | 295 | 31.92 | 5.55 | 叶绿体 |
| JrCOL28 | LOC108986213 | Chr.10:2554388..2557653 | XP_018814300.1 | JrCOL28 | 299 | 34.08 | 6.43 | 细胞核 |
| JrCOL29a-c | LOC108984383 | Chr.10:13263191..13268808 | XP_018811858.2 | JrCOL29a | 436 | 47.97 | 6.55 | 细胞质 |
| XP_035550266.1 | JrCOL29b | 436 | 47.97 | 6.55 | 细胞质 | |||
| XP_035550267.1 | JrCOL29c | 408 | 44.52 | 5.70 | 细胞核 | |||
| JrCOL30a-b | LOC109017472 | Chr.10:13911798..13913088 | XP_018855279.1 | JrCOL30a | 260 | 28.60 | 4.37 | 叶绿体 |
| XP_018855280.1 | JrCOL30b | 259 | 28.48 | 4.37 | 叶绿体 | |||
| JrCOL31a-c | LOC109000741 | Chr.10:37518173..37524266 | XP_018833263.1 | JrCOL31a | 409 | 44.45 | 5.96 | 细胞核 |
| XP_018833265.1 | JrCOL31b | 409 | 44.45 | 5.96 | 细胞核 | |||
| XP_018833266.1 | JrCOL31c | 409 | 44.45 | 5.96 | 细胞核 | |||
| JrCOL32a-d | LOC108992026 | Chr.11:8894066..8897365 | XP_018822007.1 | JrCOL32a | 194 | 21.42 | 6.43 | 叶绿体 |
| XP_018822008.1 | JrCOL32b | 192 | 21.17 | 6.23 | 叶绿体 | |||
| XP_018822009.1 | JrCOL32c | 186 | 20.48 | 6.43 | 叶绿体 | |||
| XP_018822010.1 | JrCOL32d | 184 | 20.22 | 6.23 | 叶绿体 | |||
| JrCOL33 | LOC108992144 | Chr.11:23645881..23650290 | XP_018822151.1 | JrCOL33 | 364 | 40.75 | 5.51 | 细胞核 |
| JrCOL34 | LOC108991312 | Chr.11:29101394..29103034 | XP_018821038.1 | JrCOL34 | 300 | 33.48 | 5.99 | 细胞核 |
| JrCOL35 | LOC108998881 | Chr.11:32536877..32538774 | XP_018831163.1 | JrCOL35 | 373 | 40.72 | 5.86 | 叶绿体 |
| JrCOL36 | LOC109003090 | Chr.11:33364576..33368847 | XP_018836611.1 | JrCOL36 | 409 | 45.46 | 8.94 | 细胞核 |
| JrCOL37 | LOC109005190 | Chr.11:36684126..36685787 | XP_018839540.1 | JrCOL37 | 341 | 37.58 | 6.87 | 叶绿体 |
| JrCOL38a-b | LOC108982143 | Chr.12:8537584..8542475 | XP_018808983.1 | JrCOL38a | 671 | 74.38 | 5.97 | 细胞核 |
| XP_035539295.1 | JrCOL38b | 652 | 72.23 | 6.08 | 细胞核 | |||
| JrCOL39 | LOC108992474 | Chr.12:13436453..13439714 | XP_018822597.1 | JrCOL39 | 237 | 26.10 | 4.91 | 细胞核 |
| JrCOL40a-b | LOC108981163 | Chr.12:23463905..23468842 | XP_018807797.1 | JrCOL40a | 423 | 46.11 | 4.45 | 细胞核 |
| XP_018807806.1 | JrCOL40b | 418 | 45.53 | 4.53 | 细胞核 | |||
| JrCOL41a-b | LOC108986927 | Chr.13:5686536..5688137 | XP_018815283.1 | JrCOL41a | 293 | 32.61 | 5.70 | 细胞核 |
| XP_018815285.1 | JrCOL41b | 206 | 22.72 | 5.57 | 细胞核 | |||
| JrCOL42 | LOC109008345 | Chr.13:8537826..8540900 | XP_018843947.2 | JrCOL42 | 475 | 51.86 | 5.92 | 细胞核 |
| JrCOL43a-b | LOC108992288 | Chr.13:31296667..31301250 | XP_018822350.1 | JrCOL43a | 691 | 77.57 | 6.80 | 细胞核 |
| XP_018822351.1 | JrCOL43b | 689 | 77.43 | 6.80 | 细胞核 | |||
| JrCOL44a-d | LOC109007733 | Chr.13:37236625..37242278 | XP_018843087.1 | JrCOL44a | 518 | 57.78 | 5.61 | 细胞核 |
| XP_018843095.1 | JrCOL44b | 518 | 57.78 | 5.61 | 细胞核 | |||
| XP_018843104.1 | JrCOL44c | 518 | 57.78 | 5.61 | 细胞核 | |||
| XP_018843113.1 | JrCOL44d | 518 | 57.78 | 5.61 | 细胞核 | |||
| JrCOL45 | LOC109013207 | Chr.13:38161618..38165624 | XP_018850755.1 | JrCOL45 | 287 | 32.38 | 5.25 | 细胞核 |
| JrCOL46 | LOC108996999 | Chr.14:6471961..6473893 | XP_018828617.1 | JrCOL46 | 218 | 25.99 | 5.27 | 细胞核 |
| JrCOL47 | LOC108993945 | Chr.14:10350983..10353374 | XP_018824556.1 | JrCOL47 | 251 | 28.23 | 8.27 | 细胞核 |
| JrCOL48 | LOC109013626 | Chr.15:14881124..14883084 | XP_018851315.1 | JrCOL48 | 464 | 52.39 | 5.09 | 细胞核,细胞质 |
| JrCOL49 | LOC109004589 | Chr.16:1780812..1784882 | XP_018838732.1 | JrCOL49 | 279 | 31.45 | 5.27 | 细胞核 |
| JrCOL50a-f | LOC109012501 | Chr.16:2630759..2636833 | XP_035542414.1 | JrCOL50a | 518 | 57.90 | 5.72 | 细胞核 |
| XP_035542415.1 | JrCOL50b | 492 | 54.89 | 5.87 | 细胞核 | |||
| XP_035542416.1 | JrCOL50c | 489 | 54.51 | 5.83 | 细胞核 | |||
| XP_035542417.1 | JrCOL50d | 453 | 50.39 | 5.37 | 细胞核 | |||
| XP_018849735.1 | JrCOL50e | 518 | 57.90 | 5.72 | 细胞核 | |||
| XP_018849736.1 | JrCOL50f | 518 | 57.90 | 5.72 | 细胞核 | |||
| JrCOL51 | LOC109007532 | Chr.16:6094342..6099323 | XP_018842768.2 | JrCOL51 | 709 | 78.92 | 7.28 | 细胞核 |
| JrCOL52 | LOC109008337 | Chr.16:22543220..22546364 | XP_018843939.1 | JrCOL52 | 475 | 52.12 | 6.67 | 细胞核 |
| JrCOL53a-d | LOC109006537 | Chr.16:24449761..24454424 | XP_018841385.2 | JrCOL53a | 209 | 23.20 | 6.65 | 细胞核,细胞质 |
| XP_035542160.1 | JrCOL53b | 244 | 27.19 | 7.48 | 细胞质 | |||
| XP_035542161.1 | JrCOL53c | 244 | 27.19 | 7.48 | 细胞质 | |||
| XP_035542162.1 | JrCOL53d | 209 | 23.20 | 6.65 | 细胞核 | |||
| JrCOL54a-b | LOC109006510 | Chr.16:24684660..24686424 | XP_018841344.1 | JrCOL54a | 298 | 33.48 | 6.31 | 细胞核 |
| XP_018841345.1 | JrCOL54b | 215 | 23.72 | 6.05 | 细胞核 | |||
| JrCOL55 | LOC108984240 | Scaffold_15:88..2233 | XP_018811678.1 | JrCOL55 | 170 | 18.25 | 4.08 | 细胞核 |
Table 2 COL gene family identified in Juglans regia
| 基因名称 Gene name | 基因ID Gene ID | 染色体位置 Chromosome position | 相关蛋白 Related protein | 蛋白重命名 Protein rename | 氨基酸长度 aa length | 蛋白分子量 Molecular weight/kD | 理论等电点pI | 亚细胞定位Subc- ellular localization |
|---|---|---|---|---|---|---|---|---|
| JrCOL1 | LOC109001350 | Chr.1:1256032..1258201 | XP_018834143.1 | JrCOL1 | 373 | 41.31 | 5.17 | 叶绿体 |
| JrCOL2 | LOC109001381 | Chr.1:1488506..1490928 | XP_035544637.1 | JrCOL2 | 211 | 23.40 | 5.73 | 细胞质 |
| JrCOL3 | LOC109009665 | Chr.1:3978995..3983057 | XP_018845790.2 | JrCOL3 | 350 | 39.64 | 8.72 | 细胞核 |
| JrCOL4a-d | LOC109010156 | Chr.1:18366251..18370500 | XP_035543378.1 | JrCOL4a | 428 | 47.24 | 5.27 | 细胞核 |
| XP_018846422.1 | JrCOL4b | 428 | 47.24 | 5.27 | 细胞核 | |||
| XP_018846423.1 | JrCOL4c | 428 | 47.24 | 5.27 | 细胞核 | |||
| XP_018846424.1 | JrCOL4d | 428 | 47.24 | 5.27 | 细胞核 | |||
| JrCOL5 | LOC109015440 | Chr.1:18372462..18372860 | XP_018853453.2 | JrCOL5 | 132 | 14.50 | 6.85 | 细胞核 |
| JrCOL6a-b | LOC108991987 | Chr.1:19317524..19318826 | XP_018821963.2 | JrCOL6a | 237 | 25.91 | 4.42 | 细胞核 |
| XP_018821964.2 | JrCOL6b | 236 | 25.78 | 4.42 | 细胞核 | |||
| JrCOL7a-c | LOC109000761 | Chr.1:44957519..44963229 | XP_018833298.1 | JrCOL7a | 416 | 45.11 | 5.19 | 细胞核 |
| XP_018833299.1 | JrCOL7b | 416 | 45.11 | 5.19 | 细胞核 | |||
| XP_018833303.1 | JrCOL7c | 416 | 45.11 | 5.19 | 细胞核 | |||
| JrCOL8 | LOC109013302 | Chr.2:7141021..7147381 | XP_018850883.1 | JrCOL8 | 539 | 60.48 | 6.22 | 细胞核 |
| JrCOL9 | LOC108984132 | Chr.2:11707792..11709460 | XP_018811512.1 | JrCOL9 | 277 | 31.23 | 4.94 | 细胞核 |
| JrCO10 | LOC109014521 | Chr.2:30680768..30682741 | XP_018852564.1 | JrCOL10 | 333 | 37.49 | 4.97 | 细胞核 |
| JrCOL11a-b | LOC109014085 | Chr.2:35047335..35048788 | XP_018851954.1 | JrCOL11a | 258 | 27.76 | 4.28 | 细胞核 |
| XP_018851962.1 | JrCOL11b | 255 | 27.37 | 4.25 | 细胞核 | |||
| JrCOL12a-b | LOC108999038 | Chr.3:18899526..18901860 | XP_018831379.1 | JrCOL12a | 150 | 16.21 | 4.00 | 细胞核,叶绿体 |
| XP_035544064.1 | JrCOL12b | 150 | 16.21 | 4.00 | 细胞核,叶绿体 | |||
| JrCOL13 | LOC109017893 | Chr.4:14653613..14655876 | XP_035544914.1 | JrCOL13 | 170 | 18.24 | 4.11 | 细胞核 |
| JrCOL14 | LOC108989855 | Chr.5:6297905..6299253 | XP_018819161.1 | JrCOL14 | 220 | 26.37 | 4.94 | 细胞核 |
| JrCOL15a-b | LOC109022307 | Chr.5:9188186..9197431 | XP_018860727.1 | JrCOL15a | 789 | 86.08 | 6.30 | 叶绿体 |
| XP_018860748.1 | JrCOL15b | 778 | 84.89 | 6.35 | 叶绿体 | |||
| JrCOL16 | LOC108994549 | Chr.6:15543376..15545802 | XP_018825341.2 | JrCOL16 | 444 | 48.81 | 5.88 | 细胞核 |
| JrCOL17 | LOC108998057 | Chr.6:29988891..29991120 | XP_018830038.2 | JrCOL17 | 407 | 46.32 | 5.46 | 细胞核 |
| JrCOL18 | LOC108992642 | Chr.7:12544651..12554366 | XP_018822785.2 | JrCOL18 | 685 | 75.72 | 6.71 | 细胞核 |
| JrCOL19 | LOC108983659 | Chr.7:14608063..14609540 | XP_018810913.1 | JrCOL19 | 266 | 28.49 | 8.73 | 叶绿体 |
| JrCOL20 | LOC109005469 | Chr.7:32506953..32517320 | XP_018839979.2 | JrCOL20 | 238 | 26.36 | 5.08 | 细胞核 |
| JrCOL21 | LOC109019524 | Chr.7:38617705..38622031 | XP_018857358.1 | JrCOL21 | 432 | 46.81 | 4.35 | 叶绿体 |
| JrCOL22a-b | LOC109014549 | Chr.7:49644025..49658252 | XP_035548321.1 | JrCOL22a | 780 | 85.75 | 6.42 | 叶绿体 |
| XP_035548322.1 | JrCOL22b | 778 | 85.50 | 6.42 | 叶绿体 | |||
| JrCOL23 | LOC109000996 | Chr.8:600112..601757 | XP_018833641.1 | JrCOL23 | 345 | 37.98 | 6.16 | 细胞质 |
| JrCOL24 | LOC108984911 | Chr.8:6608517..6610109 | XP_018812557.1 | JrCOL24 | 301 | 33.04 | 6.25 | 细胞核 |
| JrCOL25a-b | LOC108980667 | Chr.8:24595513..24597399 | XP_035549568.1 | JrCOL25a | 158 | 17.12 | 5.62 | 细胞质 |
| XP_035549569.1 | JrCOL25b | 117 | 12.99 | 6.49 | 细胞质 | |||
| JrCOL26 | LOC108997344 | Chr.9:19840907..19842615 | XP_018829103.1 | JrCOL26 | 331 | 37.53 | 5.83 | 细胞核 |
| JrCOL27 | LOC108985349 | Chr.9:21400481..21403221 | XP_018813169.1 | JrCOL27 | 295 | 31.92 | 5.55 | 叶绿体 |
| JrCOL28 | LOC108986213 | Chr.10:2554388..2557653 | XP_018814300.1 | JrCOL28 | 299 | 34.08 | 6.43 | 细胞核 |
| JrCOL29a-c | LOC108984383 | Chr.10:13263191..13268808 | XP_018811858.2 | JrCOL29a | 436 | 47.97 | 6.55 | 细胞质 |
| XP_035550266.1 | JrCOL29b | 436 | 47.97 | 6.55 | 细胞质 | |||
| XP_035550267.1 | JrCOL29c | 408 | 44.52 | 5.70 | 细胞核 | |||
| JrCOL30a-b | LOC109017472 | Chr.10:13911798..13913088 | XP_018855279.1 | JrCOL30a | 260 | 28.60 | 4.37 | 叶绿体 |
| XP_018855280.1 | JrCOL30b | 259 | 28.48 | 4.37 | 叶绿体 | |||
| JrCOL31a-c | LOC109000741 | Chr.10:37518173..37524266 | XP_018833263.1 | JrCOL31a | 409 | 44.45 | 5.96 | 细胞核 |
| XP_018833265.1 | JrCOL31b | 409 | 44.45 | 5.96 | 细胞核 | |||
| XP_018833266.1 | JrCOL31c | 409 | 44.45 | 5.96 | 细胞核 | |||
| JrCOL32a-d | LOC108992026 | Chr.11:8894066..8897365 | XP_018822007.1 | JrCOL32a | 194 | 21.42 | 6.43 | 叶绿体 |
| XP_018822008.1 | JrCOL32b | 192 | 21.17 | 6.23 | 叶绿体 | |||
| XP_018822009.1 | JrCOL32c | 186 | 20.48 | 6.43 | 叶绿体 | |||
| XP_018822010.1 | JrCOL32d | 184 | 20.22 | 6.23 | 叶绿体 | |||
| JrCOL33 | LOC108992144 | Chr.11:23645881..23650290 | XP_018822151.1 | JrCOL33 | 364 | 40.75 | 5.51 | 细胞核 |
| JrCOL34 | LOC108991312 | Chr.11:29101394..29103034 | XP_018821038.1 | JrCOL34 | 300 | 33.48 | 5.99 | 细胞核 |
| JrCOL35 | LOC108998881 | Chr.11:32536877..32538774 | XP_018831163.1 | JrCOL35 | 373 | 40.72 | 5.86 | 叶绿体 |
| JrCOL36 | LOC109003090 | Chr.11:33364576..33368847 | XP_018836611.1 | JrCOL36 | 409 | 45.46 | 8.94 | 细胞核 |
| JrCOL37 | LOC109005190 | Chr.11:36684126..36685787 | XP_018839540.1 | JrCOL37 | 341 | 37.58 | 6.87 | 叶绿体 |
| JrCOL38a-b | LOC108982143 | Chr.12:8537584..8542475 | XP_018808983.1 | JrCOL38a | 671 | 74.38 | 5.97 | 细胞核 |
| XP_035539295.1 | JrCOL38b | 652 | 72.23 | 6.08 | 细胞核 | |||
| JrCOL39 | LOC108992474 | Chr.12:13436453..13439714 | XP_018822597.1 | JrCOL39 | 237 | 26.10 | 4.91 | 细胞核 |
| JrCOL40a-b | LOC108981163 | Chr.12:23463905..23468842 | XP_018807797.1 | JrCOL40a | 423 | 46.11 | 4.45 | 细胞核 |
| XP_018807806.1 | JrCOL40b | 418 | 45.53 | 4.53 | 细胞核 | |||
| JrCOL41a-b | LOC108986927 | Chr.13:5686536..5688137 | XP_018815283.1 | JrCOL41a | 293 | 32.61 | 5.70 | 细胞核 |
| XP_018815285.1 | JrCOL41b | 206 | 22.72 | 5.57 | 细胞核 | |||
| JrCOL42 | LOC109008345 | Chr.13:8537826..8540900 | XP_018843947.2 | JrCOL42 | 475 | 51.86 | 5.92 | 细胞核 |
| JrCOL43a-b | LOC108992288 | Chr.13:31296667..31301250 | XP_018822350.1 | JrCOL43a | 691 | 77.57 | 6.80 | 细胞核 |
| XP_018822351.1 | JrCOL43b | 689 | 77.43 | 6.80 | 细胞核 | |||
| JrCOL44a-d | LOC109007733 | Chr.13:37236625..37242278 | XP_018843087.1 | JrCOL44a | 518 | 57.78 | 5.61 | 细胞核 |
| XP_018843095.1 | JrCOL44b | 518 | 57.78 | 5.61 | 细胞核 | |||
| XP_018843104.1 | JrCOL44c | 518 | 57.78 | 5.61 | 细胞核 | |||
| XP_018843113.1 | JrCOL44d | 518 | 57.78 | 5.61 | 细胞核 | |||
| JrCOL45 | LOC109013207 | Chr.13:38161618..38165624 | XP_018850755.1 | JrCOL45 | 287 | 32.38 | 5.25 | 细胞核 |
| JrCOL46 | LOC108996999 | Chr.14:6471961..6473893 | XP_018828617.1 | JrCOL46 | 218 | 25.99 | 5.27 | 细胞核 |
| JrCOL47 | LOC108993945 | Chr.14:10350983..10353374 | XP_018824556.1 | JrCOL47 | 251 | 28.23 | 8.27 | 细胞核 |
| JrCOL48 | LOC109013626 | Chr.15:14881124..14883084 | XP_018851315.1 | JrCOL48 | 464 | 52.39 | 5.09 | 细胞核,细胞质 |
| JrCOL49 | LOC109004589 | Chr.16:1780812..1784882 | XP_018838732.1 | JrCOL49 | 279 | 31.45 | 5.27 | 细胞核 |
| JrCOL50a-f | LOC109012501 | Chr.16:2630759..2636833 | XP_035542414.1 | JrCOL50a | 518 | 57.90 | 5.72 | 细胞核 |
| XP_035542415.1 | JrCOL50b | 492 | 54.89 | 5.87 | 细胞核 | |||
| XP_035542416.1 | JrCOL50c | 489 | 54.51 | 5.83 | 细胞核 | |||
| XP_035542417.1 | JrCOL50d | 453 | 50.39 | 5.37 | 细胞核 | |||
| XP_018849735.1 | JrCOL50e | 518 | 57.90 | 5.72 | 细胞核 | |||
| XP_018849736.1 | JrCOL50f | 518 | 57.90 | 5.72 | 细胞核 | |||
| JrCOL51 | LOC109007532 | Chr.16:6094342..6099323 | XP_018842768.2 | JrCOL51 | 709 | 78.92 | 7.28 | 细胞核 |
| JrCOL52 | LOC109008337 | Chr.16:22543220..22546364 | XP_018843939.1 | JrCOL52 | 475 | 52.12 | 6.67 | 细胞核 |
| JrCOL53a-d | LOC109006537 | Chr.16:24449761..24454424 | XP_018841385.2 | JrCOL53a | 209 | 23.20 | 6.65 | 细胞核,细胞质 |
| XP_035542160.1 | JrCOL53b | 244 | 27.19 | 7.48 | 细胞质 | |||
| XP_035542161.1 | JrCOL53c | 244 | 27.19 | 7.48 | 细胞质 | |||
| XP_035542162.1 | JrCOL53d | 209 | 23.20 | 6.65 | 细胞核 | |||
| JrCOL54a-b | LOC109006510 | Chr.16:24684660..24686424 | XP_018841344.1 | JrCOL54a | 298 | 33.48 | 6.31 | 细胞核 |
| XP_018841345.1 | JrCOL54b | 215 | 23.72 | 6.05 | 细胞核 | |||
| JrCOL55 | LOC108984240 | Scaffold_15:88..2233 | XP_018811678.1 | JrCOL55 | 170 | 18.25 | 4.08 | 细胞核 |
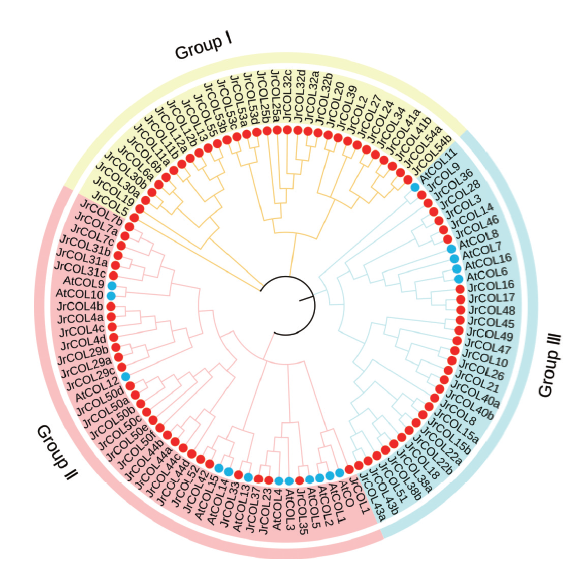
Fig. 1 Phylogenetic relationship of the COL gene family of Arabidopsis and walnut(Juglans regia) The red solid dot represents walnut COL protein,blue solid dot represents Arabidopsis COL protein
| 顺式作用元件 Cis-element | 基因数目 Gene number | 序列 Sequence | 功能 Function | 顺式作用元件类型 Cis-element type |
|---|---|---|---|---|
| ARE | 46 | AAACCA | 厌氧诱导所必需的顺式作用调控元件 cis-elements for anaerobic induction | 胁迫相关响应元件 Abiotic and biotic stresses |
| DRE core | 10 | GCCGAC | 脱水响应元件Dehydration-responsive element | |
| LTR | 18 | CCGAAA | 参与低温反应的顺式作用元件 Low-temperature responsive cis-element | |
| MBS | 17 | CAACTG | MYB结合位点参与干旱诱导 MYB binding site involved in drought induction | |
| MYB | 50 | CAACAG | MYB响应元件MYB-responsive element | |
| MYC | 53 | CAATTG | MYC响应元件MYC-responsive element | |
| STRE | 40 | AGGGG | 胁迫响应元件Stress-responsive element | |
| TC-rich repeat | 10 | GTTTTCTTAC | 参与防御和胁迫反应的顺式作用元素 Defense and stress-responsive element | |
| WUN-motif | 21 | AAATTACT | 创伤响应元件Mechanical wounding element | |
| ABRE | 43 | ACGTG | 脱落酸反应中的顺式作用元件ABA-responsive element | 激素相关响应元件 Phytohormone responsive |
| ERE | 32 | ATTTTAAA | 乙烯响应元件Ethephon-responsive element | |
| TGA-element | 14 | AACGAC | 生长素响应元件Auxin-responsive element | |
| TCA-element | 16 | CAGAAAAGGA | 参与水杨酸反应的顺式作用元件 Salicylic-acid-responsive element | |
| TGACG-motif | 33 | TGACG | 参与MeJA反应的顺调控元件MeJA-responsive element | |
| CGTCA-motif | 34 | CGTCA | 参与MeJA反应的顺调控元件MeJA-responsive element | |
| P-box | 16 | CCTTTTG | 赤霉素响应元件Gibberellin-responsive element | |
| TATC-box | 9 | TATCCCA | 赤霉素响应元件Gibberellin-responsive element | |
| Box 4 | 42 | ATTAAT | 与光反应有关的保守DNA模块的一部分 Part of a conserved DNA module involved in light responsiveness | 生长发育相关响应元件 Plant growth and development |
| CAT-box | 13 | GCCACT | 与分生组织表达相关的调控元件 Involved in Meristem expression element | |
| GA-motif | 9 | ATAGATAA | 光响应模块的一部分Part of a light-responsive module | |
| GATA-motif | 21 | AAGGATAAGG | 光响应元件的一部分Part of a light-responsive element | |
| G-box | 49 | CACGTC | 参与光响应的调控元件Involved in light-responsive element | |
| GT1-motif | 23 | GGTTAAT | 光响应元件Light-responsive element |
Table 3 Prediction of cis-element in the JrCOL promoter
| 顺式作用元件 Cis-element | 基因数目 Gene number | 序列 Sequence | 功能 Function | 顺式作用元件类型 Cis-element type |
|---|---|---|---|---|
| ARE | 46 | AAACCA | 厌氧诱导所必需的顺式作用调控元件 cis-elements for anaerobic induction | 胁迫相关响应元件 Abiotic and biotic stresses |
| DRE core | 10 | GCCGAC | 脱水响应元件Dehydration-responsive element | |
| LTR | 18 | CCGAAA | 参与低温反应的顺式作用元件 Low-temperature responsive cis-element | |
| MBS | 17 | CAACTG | MYB结合位点参与干旱诱导 MYB binding site involved in drought induction | |
| MYB | 50 | CAACAG | MYB响应元件MYB-responsive element | |
| MYC | 53 | CAATTG | MYC响应元件MYC-responsive element | |
| STRE | 40 | AGGGG | 胁迫响应元件Stress-responsive element | |
| TC-rich repeat | 10 | GTTTTCTTAC | 参与防御和胁迫反应的顺式作用元素 Defense and stress-responsive element | |
| WUN-motif | 21 | AAATTACT | 创伤响应元件Mechanical wounding element | |
| ABRE | 43 | ACGTG | 脱落酸反应中的顺式作用元件ABA-responsive element | 激素相关响应元件 Phytohormone responsive |
| ERE | 32 | ATTTTAAA | 乙烯响应元件Ethephon-responsive element | |
| TGA-element | 14 | AACGAC | 生长素响应元件Auxin-responsive element | |
| TCA-element | 16 | CAGAAAAGGA | 参与水杨酸反应的顺式作用元件 Salicylic-acid-responsive element | |
| TGACG-motif | 33 | TGACG | 参与MeJA反应的顺调控元件MeJA-responsive element | |
| CGTCA-motif | 34 | CGTCA | 参与MeJA反应的顺调控元件MeJA-responsive element | |
| P-box | 16 | CCTTTTG | 赤霉素响应元件Gibberellin-responsive element | |
| TATC-box | 9 | TATCCCA | 赤霉素响应元件Gibberellin-responsive element | |
| Box 4 | 42 | ATTAAT | 与光反应有关的保守DNA模块的一部分 Part of a conserved DNA module involved in light responsiveness | 生长发育相关响应元件 Plant growth and development |
| CAT-box | 13 | GCCACT | 与分生组织表达相关的调控元件 Involved in Meristem expression element | |
| GA-motif | 9 | ATAGATAA | 光响应模块的一部分Part of a light-responsive module | |
| GATA-motif | 21 | AAGGATAAGG | 光响应元件的一部分Part of a light-responsive element | |
| G-box | 49 | CACGTC | 参与光响应的调控元件Involved in light-responsive element | |
| GT1-motif | 23 | GGTTAAT | 光响应元件Light-responsive element |
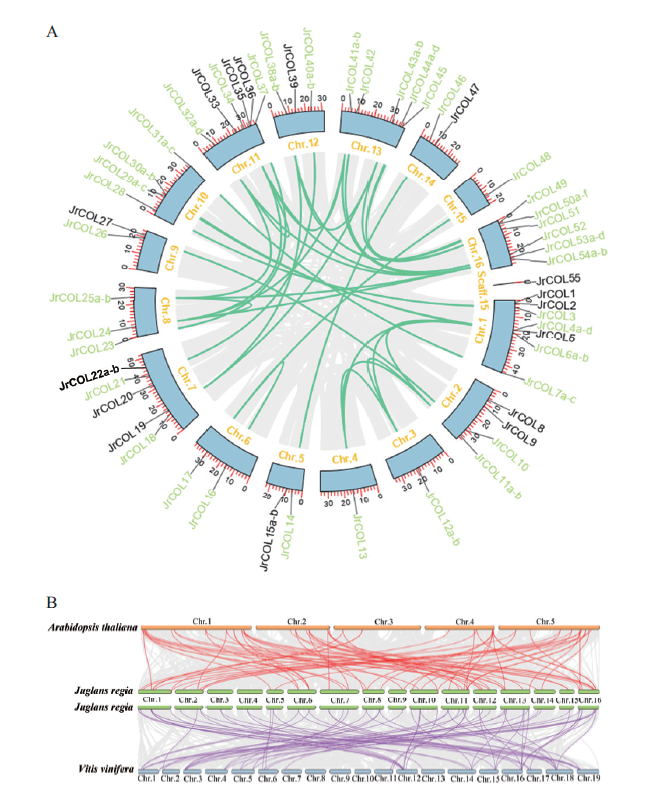
Fig. 3 Duplication models of the COL genes in walnut A:Synteny analysis of JrCOL genes. Gray lines indicate all synteny blocks in the walnut genome,whereas segmental duplicated homologous genes are indicated with green line. B:Synteny analysis of COL genes between Arabidopsis,walnut and grape. The gray lines in the background indicate the collinear blocks within walnut and other plant genomes,while the red and purple lines highlight the syntenic COL gene pairs
| 同源基因对 Homologous gene pair | Ka | Ks | Ka/Ks |
|---|---|---|---|
| JrCOL3 & JrCOL28 | 0.164 | 0.426 | 0.386 |
| JrCOL4a-d & JrCOL29a-c | 0.120 | 0.310 | 0.388 |
| JrCOL6a-b & JrCOL11a-b | 0.371 | 1.753 | 0.212 |
| JrCOL6a-b & JrCOL12a-b | —— | —— | —— |
| JrCOL6a-b & JrCOL13 | —— | —— | —— |
| JrCOL6a-b & JrCOL30a-b | 0.163 | 0.405 | 0.402 |
| JrCOL7a-c & JrCOL31a-c | 0.082 | 0.316 | 0.258 |
| JrCOL10 & JrCOL26 | 0.105 | 0.349 | 0.301 |
| JrCOL11a-b & JrCOL12a-b | —— | —— | —— |
| JrCOL11a-b & JrCOL13 | —— | —— | —— |
| JrCOL12a-b & JrCOL13 | 0.136 | 0.690 | 0.197 |
| JrCOL14 & JrCOL46 | 0.095 | 0.467 | 0.203 |
| JrCOL16 & JrCOL17 | 0.315 | 2.218 | 0.142 |
| JrCOL17 & JrCOL48 | 0.158 | 0.352 | 0.449 |
| JrCOL18 & JrCOL38a-b | 0.164 | 0.339 | 0.484 |
| JrCOL18 & JrCOL43a-b | 0.445 | 1.224 | 0.364 |
| JrCOL21 & JrCOL40a-b | 0.081 | 0.309 | 0.263 |
| JrCOL23 & JrCOL37 | 0.111 | 0.506 | 0.220 |
| JrCOL24 & JrCOL25a-b | —— | —— | —— |
| JrCOL24 & JrCOL34 | 0.123 | 0.347 | 0.354 |
| JrCOL24 & JrCOL41a-b | 0.351 | 5.257 | 0.067 |
| JrCOL24 & JrCOL54a-b | 0.338 | 2.787 | 0.121 |
| JrCOL25a-b & JrCOL32a-d | 0.061 | 0.341 | 0.179 |
| JrCOL25a-b & JrCOL53a-d | 0.164 | 1.270 | 0.129 |
| JrCOL32a-d & JrCOL34 | 0.627 | 3.152 | 0.199 |
| JrCOL32a-d & JrCOL53a-d | 0.223 | 1.214 | 0.184 |
| JrCOL34 & JrCOL41a-b | 0.385 | 1.660 | 0.232 |
| JrCOL34 & JrCOL54a-b | 0.342 | 1.956 | 0.175 |
| JrCOL38a-b & JrCOL43a-b | 0.418 | 1.346 | 0.310 |
| JrCOL38a-b & JrCOL51 | 0.429 | 1.279 | 0.336 |
| JrCOL41a-b & JrCOL54a-b | 0.163 | 0.466 | 0.349 |
| JrCOL42 & JrCOL52 | 0.111 | 0.433 | 0.256 |
| JrCOL43a-b & JrCOL51 | 0.136 | 0.329 | 0.413 |
| JrCOL44a-d & JrCOL50a-f | 0.140 | 0.316 | 0.442 |
| JrCOL45 & JrCOL49 | 0.069 | 0.426 | 0.161 |
Table 4 Analysis of duplication time reflected from Ka/Ks values of JrCOL gene family
| 同源基因对 Homologous gene pair | Ka | Ks | Ka/Ks |
|---|---|---|---|
| JrCOL3 & JrCOL28 | 0.164 | 0.426 | 0.386 |
| JrCOL4a-d & JrCOL29a-c | 0.120 | 0.310 | 0.388 |
| JrCOL6a-b & JrCOL11a-b | 0.371 | 1.753 | 0.212 |
| JrCOL6a-b & JrCOL12a-b | —— | —— | —— |
| JrCOL6a-b & JrCOL13 | —— | —— | —— |
| JrCOL6a-b & JrCOL30a-b | 0.163 | 0.405 | 0.402 |
| JrCOL7a-c & JrCOL31a-c | 0.082 | 0.316 | 0.258 |
| JrCOL10 & JrCOL26 | 0.105 | 0.349 | 0.301 |
| JrCOL11a-b & JrCOL12a-b | —— | —— | —— |
| JrCOL11a-b & JrCOL13 | —— | —— | —— |
| JrCOL12a-b & JrCOL13 | 0.136 | 0.690 | 0.197 |
| JrCOL14 & JrCOL46 | 0.095 | 0.467 | 0.203 |
| JrCOL16 & JrCOL17 | 0.315 | 2.218 | 0.142 |
| JrCOL17 & JrCOL48 | 0.158 | 0.352 | 0.449 |
| JrCOL18 & JrCOL38a-b | 0.164 | 0.339 | 0.484 |
| JrCOL18 & JrCOL43a-b | 0.445 | 1.224 | 0.364 |
| JrCOL21 & JrCOL40a-b | 0.081 | 0.309 | 0.263 |
| JrCOL23 & JrCOL37 | 0.111 | 0.506 | 0.220 |
| JrCOL24 & JrCOL25a-b | —— | —— | —— |
| JrCOL24 & JrCOL34 | 0.123 | 0.347 | 0.354 |
| JrCOL24 & JrCOL41a-b | 0.351 | 5.257 | 0.067 |
| JrCOL24 & JrCOL54a-b | 0.338 | 2.787 | 0.121 |
| JrCOL25a-b & JrCOL32a-d | 0.061 | 0.341 | 0.179 |
| JrCOL25a-b & JrCOL53a-d | 0.164 | 1.270 | 0.129 |
| JrCOL32a-d & JrCOL34 | 0.627 | 3.152 | 0.199 |
| JrCOL32a-d & JrCOL53a-d | 0.223 | 1.214 | 0.184 |
| JrCOL34 & JrCOL41a-b | 0.385 | 1.660 | 0.232 |
| JrCOL34 & JrCOL54a-b | 0.342 | 1.956 | 0.175 |
| JrCOL38a-b & JrCOL43a-b | 0.418 | 1.346 | 0.310 |
| JrCOL38a-b & JrCOL51 | 0.429 | 1.279 | 0.336 |
| JrCOL41a-b & JrCOL54a-b | 0.163 | 0.466 | 0.349 |
| JrCOL42 & JrCOL52 | 0.111 | 0.433 | 0.256 |
| JrCOL43a-b & JrCOL51 | 0.136 | 0.329 | 0.413 |
| JrCOL44a-d & JrCOL50a-f | 0.140 | 0.316 | 0.442 |
| JrCOL45 & JrCOL49 | 0.069 | 0.426 | 0.161 |
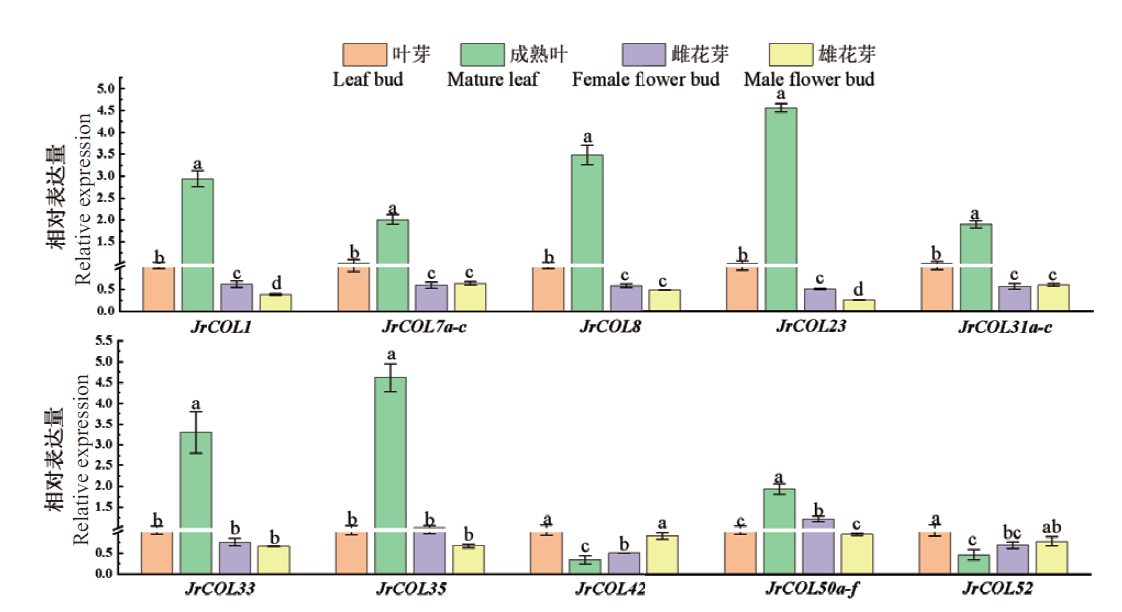
Fig. 5 Relative expressions of JrCOL genes in Juglans regia cv. Xinxin No. 2 in different tissues Different lowercase letters indicate significant differences between treatments(P<0.05). The same below
| [1] |
Milyaev A, Kofler J, Klaiber I, et al. Toward systematic understanding of flower bud induction in apple:a multi-omics approach[J]. Front Plant Sci, 2021, 12:604810.
doi: 10.3389/fpls.2021.604810 URL |
| [2] |
Griffiths S, Dunford RP, Coupland G, et al. The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis[J]. Plant Physiol, 2003, 131(4):1855-1867.
pmid: 12692345 |
| [3] |
Imaizumi T, Schultz TF, Harmon FG, et al. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis[J]. Science, 2005, 309(5732):293-297.
pmid: 16002617 |
| [4] |
Putterill J, Robson F, Lee K, et al. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors[J]. Cell, 1995, 80(6):847-857.
pmid: 7697715 |
| [5] |
Hassidim M, Harir Y, Yakir E, et al. Over-expression of CONSTANS-LIKE 5 can induce flowering in short-day grown Arabidopsis[J]. Planta, 2009, 230(3):481-491.
doi: 10.1007/s00425-009-0958-7 pmid: 19504268 |
| [6] |
Cheng XF, Wang ZY. Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana[J]. Plant J, 2005, 43(5):758-768.
doi: 10.1111/j.1365-313X.2005.02491.x URL |
| [7] |
Takase T, Kakikubo Y, Nakasone A, et al. Characterization and transgenic study of CONSTANS-LIKE8(COL8)gene in Arabidopsis thaliana:expression of 35S:COL8 delays flowering under long-day conditions[J]. Plant Biotechnol, 2011, 28(5):439-446.
doi: 10.5511/plantbiotechnology.11.0823b URL |
| [8] |
Kim SK, Yun CH, Lee JH, et al. OsCO3, a CONSTANS-LIKE gene, controls flowering by negatively regulating the expression of FT-like genes under SD conditions in rice[J]. Planta, 2008, 228(2):355-365.
doi: 10.1007/s00425-008-0742-0 URL |
| [9] |
Sheng PK, Wu FQ, Tan JJ, et al. A CONSTANS-like transcriptional activator, OsCOL13, functions as a negative regulator of flowering downstream of OsphyB and upstream of Ehd1 in rice[J]. Plant Mol Biol, 2016, 92(1-2):209-222.
doi: 10.1007/s11103-016-0506-3 pmid: 27405463 |
| [10] |
Tan JJ, Jin MN, Wang JC, et al. OsCOL10, a CONSTANS-like gene, functions as a flowering time repressor downstream of Ghd7 in rice[J]. Plant Cell Physiol, 2016, 57(4):798-812.
doi: 10.1093/pcp/pcw025 URL |
| [11] |
Campoli C, Drosse B, Searle I, et al. Functional characterisation of HvCO1, the barley(Hordeum vulgare)flowering time ortholog of CONSTANS[J]. Plant J, 2012, 69(5):868-880.
doi: 10.1111/j.1365-313X.2011.04839.x URL |
| [12] |
Kim SJ, Moon J, Lee I, et al. Molecular cloning and expression analysis of a CONSTANS homologue, PnCOL1, from Pharbitis nil[J]. J Exp Bot, 2003, 54(389):1879-1887.
doi: 10.1093/jxb/erg217 URL |
| [13] |
Ledger S, Strayer C, Ashton F, et al. Analysis of the function of two circadian-regulated CONSTANS-LIKE genes[J]. Plant J, 2001, 26(1):15-22.
pmid: 11359606 |
| [14] |
Liu L, Ma J, Han Y, et al. The isolation and analysis of a soybean CO Homologue GmCOL10[J]. Russ J Plant Physiol, 2011, 58(2):330-336.
doi: 10.1134/S1021443711020105 URL |
| [15] |
Datta S, Hettiarachchi GHCM, Deng XW, et al. Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth[J]. Plant Cell, 2006, 18(1):70-84.
doi: 10.1105/tpc.105.038182 URL |
| [16] |
Min JH, Chung JS, Lee KH, et al. The CONSTANS-like 4 transcription factor, AtCOL4, positively regulates abiotic stress tolerance through an abscisic acid-dependent manner in Arabidopsis[J]. J Integr Plant Biol, 2015, 57(3):313-324.
doi: 10.1111/jipb.12246 URL |
| [17] |
González-Schain ND, Suárez-López P. CONSTANS delays flowering and affects tuber yield in potato[J]. Biol Plant, 2008, 52(2):251-258.
doi: 10.1007/s10535-008-0054-z URL |
| [18] |
Chen J, Chen JY, Wang JN, et al. Molecular characterization and expression profiles of MaCOL1, a CONSTANS-like gene in banana fruit[J]. Gene, 2012, 496(2):110-117.
doi: 10.1016/j.gene.2012.01.008 URL |
| [19] | Deng XD, Fan XZ, Li P, et al. A photoperiod-regulating gene CONSTANS is correlated to lipid biosynthesis in Chlamydomonas reinhardtii[J]. Biomed Res Int, 2015, 2015:715020. |
| [20] |
Almada R, Cabrera N, Casaretto JA, et al. VvCO and VvCOL1, two CONSTANS homologous genes, are regulated during flower induction and dormancy in grapevine buds[J]. Plant Cell Rep, 2009, 28(8):1193-1203.
doi: 10.1007/s00299-009-0720-4 pmid: 19495771 |
| [21] |
Hayes D, Angove MJ, Tucci J, et al. Walnuts(Juglans regia)chemical composition and research in human health[J]. Crit Rev Food Sci Nutr, 2016, 56(8):1231-1241.
doi: 10.1080/10408398.2012.760516 URL |
| [22] |
Robson F, Costa MM, Hepworth SR, et al. Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants[J]. Plant J, 2001, 28(6):619-631.
pmid: 11851908 |
| [23] |
Wu FQ, Price BW, Haider W, et al. Functional and evolutionary characterization of the CONSTANS gene family in short-day photoperiodic flowering in soybean[J]. PLoS One, 2014, 9(1):e85754.
doi: 10.1371/journal.pone.0085754 URL |
| [24] |
Song XM, Duan WK, Huang ZN, et al. Comprehensive analysis of the flowering genes in Chinese cabbage and examination of evolutionary pattern of CO-like genes in plant kingdom[J]. Sci Rep, 2015, 5:14631.
doi: 10.1038/srep14631 URL |
| [25] |
Fu JX, Yang LW, Dai SL. Identification and characterization of the CONSTANS-like gene family in the short-day plant Chrysanthemum lavandulifolium[J]. Mol Genet Genom, 2015, 290(3):1039-1054.
doi: 10.1007/s00438-014-0977-3 URL |
| [26] |
Hu TH, Wei QZ, Wang WH, et al. Genome-wide identification and characterization of CONSTANS-like gene family in radish(Raphanus sativus)[J]. PLoS One, 2018, 13(9):e0204137.
doi: 10.1371/journal.pone.0204137 URL |
| [27] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method[J]. Methods, 2001, 25(4):402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [28] |
Quan SW, Niu JX, Zhou L, et al. Stages identifying and transcriptome profiling of the floral transition in Juglans regia[J]. Sci Rep, 2019, 9(1):7092.
doi: 10.1038/s41598-019-43582-z URL |
| [29] |
Li J, Gao K, Yang XY, et al. Identification and characterization of the CONSTANS-like gene family and its expression profiling under light treatment in Populus[J]. Int J Biol Macromol, 2020, 161:999-1010.
doi: 10.1016/j.ijbiomac.2020.06.056 URL |
| [30] |
Song NN, Xu ZL, Wang J, et al. Genome-wide analysis of maize CONSTANS-LIKE gene family and expression profiling under light/dark and abscisic acid treatment[J]. Gene, 2018, 673:1-11.
doi: 10.1016/j.gene.2018.06.032 URL |
| [31] |
Kinoshita A, Richter R. Genetic and molecular basis of floral induction in Arabidopsis thaliana[J]. J Exp Bot, 2020, 71(9):2490-2504.
doi: 10.1093/jxb/eraa057 pmid: 32067033 |
| [32] |
Tan JJ, Wu FQ, Wan JM. Flowering time regulation by the CONSTANS-like gene OsCOL10[J]. Plant Signal Behav, 2017, 12(1):e1267893.
doi: 10.1080/15592324.2016.1267893 URL |
| [33] |
Xu F, Li T, Xu PB, et al. DELLA proteins physically interact with CONSTANS to regulate flowering under long days in Arabidopsis[J]. FEBS Lett, 2016, 590(4):541-549.
doi: 10.1002/1873-3468.12076 URL |
| [34] |
Stevens ME, Woeste KE, Pijut PM. Localized gene expression changes during adventitious root formation in black walnut(Juglans nigra L.)[J]. Tree Physiol, 2018, 38(6):877-894.
doi: 10.1093/treephys/tpx175 pmid: 29378021 |
| [35] |
Liu H, Dong SY, Sun DY, et al. CONSTANS-like 9(OsCOL9)interacts with receptor for activated C-kinase 1(OsRACK1)to regulate blast resistance through salicylic acid and ethylene signaling pathways[J]. PLoS One, 2016, 11(11):e0166249.
doi: 10.1371/journal.pone.0166249 URL |
| [36] |
Hayama R, Yokoi S, Tamaki S, et al. Adaptation of photoperiodic control pathways produces short-day flowering in rice[J]. Nature, 2003, 422(6933):719-722.
doi: 10.1038/nature01549 URL |
| [37] |
Wigge PA, Kim MC, Jaeger KE, et al. Integration of spatial and temporal information during floral induction in Arabidopsis[J]. Science, 2005, 309(5737):1056-1059.
doi: 10.1126/science.1114358 URL |
| [38] |
Wang L, Xue JY, Dai WN, et al. Genome-wide identification, phylogenetic analysis, and expression profiling of CONSTANS-like(COL)genes in Vitis vinifera[J]. J Plant Growth Regul, 2019, 38(2):631-643.
doi: 10.1007/s00344-018-9878-8 URL |
| [39] |
Wang P, Liu Z, Cao P, et al. PbCOL8 is a clock-regulated flowering time repressor in pear[J]. Tree Genet Genomes, 2017, 13(5):1-14.
doi: 10.1007/s11295-016-1081-0 URL |
| [40] |
Yamaguchi A, Kobayashi Y, Goto K, et al. TWIN SISTER OF FT(TSF)acts as a floral pathway integrator redundantly with FT[J]. Plant Cell Physiol, 2005, 46(8):1175-1189.
pmid: 15951566 |
| [41] |
Valverde F, Mouradov A, Soppe W, et al. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering[J]. Science, 2004, 303(5660):1003-1006.
pmid: 14963328 |
| [1] | YANG Zhi-xiao, HOU Qian, LIU Guo-quan, LU Zhi-gang, CAO Yi, GOU Jian-yu, WANG Yi, LIN Ying-chao. Responses of Rubisco and Rubisco Activase in Different Resistant Tobacco Strains to Brown Spot Stress [J]. Biotechnology Bulletin, 2023, 39(9): 202-212. |
| [2] | CHEN Zhong-yuan, WANG Yu-hong, DAI Wei-jun, ZHANG Yan-min, YE Qian, LIU Xu-ping, TAN Wen-Song, ZHAO Liang. Mechanism Investigation of Ferric Ammonium Citrate on Transfection for Suspended HEK293 Cells [J]. Biotechnology Bulletin, 2023, 39(9): 311-318. |
| [3] | ZHANG Lu-yang, HAN Wen-long, XU Xiao-wen, YAO Jian, LI Fang-fang, TIAN Xiao-yuan, ZHANG Zhi-qiang. Identification and Expression Analysis of the Tobacco TCP Gene Family [J]. Biotechnology Bulletin, 2023, 39(6): 248-258. |
| [4] | LI Zhi-qi, YUAN Yue, MIAO Rong-qing, PANG Qiu-ying, ZHANG Ai-qin. Melatonin Contents in Eutrema salsugineum and Arabidopsis thaliana Under Salt Stress, and Expression Pattern Analysis of Synthesis Related Genes [J]. Biotechnology Bulletin, 2023, 39(5): 142-151. |
| [5] | LIU Kui, LI Xing-fen, YANG Pei-xin, ZHONG Zhao-chen, CAO Yi-bo, ZHANG Ling-yun. Functional Study and Validation of Transcriptional Coactivator PwMBF1c in Picea wilsonii [J]. Biotechnology Bulletin, 2023, 39(5): 205-216. |
| [6] | LAI Rui-lian, FENG Xin, GAO Min-xia, LU Yu-dan, LIU Xiao-chi, WU Ru-jian, CHEN Yi-ting. Genome-wide Identification of Catalase Family Genes and Expression Analysis in Kiwifruit [J]. Biotechnology Bulletin, 2023, 39(4): 136-147. |
| [7] | GUO San-bao, SONG Mei-ling, LI Ling-xin, YAO Zi-zhao, GUI Ming-ming, HUANG Sheng-he. Cloning and Analysis of Chalcone Synthase Gene and Its Promoter from Euphorbia maculata [J]. Biotechnology Bulletin, 2023, 39(4): 148-156. |
| [8] | CHEN Qiang, ZHOU Ming-kang, SONG Jia-min, ZHANG Chong, WU Long-kun. Identification and Analysis of LBD Gene Family and Expression Analysis of Fruit Development in Cucumis melo [J]. Biotechnology Bulletin, 2023, 39(3): 176-183. |
| [9] | YAO Xiao-wen, LIANG Xiao, CHEN Qing, WU Chun-ling, LIU Ying, LIU Xiao-qiang, SHUI Jun, QIAO Yang, MAO Yi-ming, CHEN Yin-hua, ZHANG Yin-dong. Study on the Expression Pattern of Genes in Lignin Biosynthesis Pathway of Cassava Resisting to Tetranychus urticae [J]. Biotechnology Bulletin, 2023, 39(2): 161-171. |
| [10] | LI Yan-xia, WANG Jin-peng, FENG Fen, BAO Bin-wu, DONG Yi-wen, WANG Xing-ping, LUORENG Zhuo-ma. Effects of Escherichia coli Dairy Cow Mastitis on the Expressions of Milk-producing Trait Related Genes [J]. Biotechnology Bulletin, 2023, 39(2): 274-282. |
| [11] | FENG Ce-ting, JIANG Lyu, LIU Xing-ying, LUO Le, PAN Hui-tang, ZHANG Qi-xiang, YU Chao. Identification of the NAC Gene Family in Rosa persica and Response Analysis Under Drought Stress [J]. Biotechnology Bulletin, 2023, 39(11): 283-296. |
| [12] | JIANG Nan, SHI Yang, ZHAO Zhi-hui, LI Bin, ZHAO Yi-hui, YANG Jun-biao, YAN Jia-ming, JIN Yu-fan, CHEN Ji, HUANG Jin. Expression and Functional Analysis of OsPT1 Gene in Rice Under Cadmium Stress [J]. Biotechnology Bulletin, 2023, 39(1): 166-174. |
| [13] | DUAN Min-jie, LI Yi-fei, YANG Xiao-miao, WANG Chun-ping, HUANG Qi-zhong, HUANG Ren-zhong, ZHANG Shi-cai. Identification of Zinc Finger Protein DnaJ-Like Gene Family in Capsicum annuum and Its Expression Analysis Responses to High Temperature Stress [J]. Biotechnology Bulletin, 2023, 39(1): 187-198. |
| [14] | GUO Bin-hui, SONG Li. Transcription of Ethylene Biosynthesis and Signaling Associated Genes in Response to Heterodera glycine Infection [J]. Biotechnology Bulletin, 2022, 38(8): 150-158. |
| [15] | ZHANG Miao, YANG Lu-lu, JIA Yan-long, WANG Tian-yun. Research Progress in the Roles of DNA and Histone Methylations in Epigenetic Regulation [J]. Biotechnology Bulletin, 2022, 38(7): 23-30. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||