








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (1): 224-231.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0461
Previous Articles Next Articles
YU Xiao-ling1,2( ), LI Wen-bin1,2, LI Zhi-bo1, RUAN Meng-bin1,2(
), LI Wen-bin1,2, LI Zhi-bo1, RUAN Meng-bin1,2( )
)
Received:2022-04-15
Online:2023-01-26
Published:2023-02-02
Contact:
RUAN Meng-bin
E-mail:yuxiaoling@itbb.org.cn;ruanmengbin@itbb.org.cn
YU Xiao-ling, LI Wen-bin, LI Zhi-bo, RUAN Meng-bin. Cold Resistance Function Analysis of Cassava MeMYC2.2[J]. Biotechnology Bulletin, 2023, 39(1): 224-231.
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 引物用途 Primer usage |
|---|---|---|
| MeMYC 2.2-F | GCTCTAGAATGAATCTGT- GGACGGACGA | 基因克隆 Gene cloning |
| MeMYC 2.2-R | GTGGATCCGGAGTCACCA- ACTTTGGTTG | |
| MeActin-qF | TGATGAGTCTGGTCCATCCA | 内参基因 |
| MeActin-qR | CCTCCTACGACCCAATCTCA | Reference gene |
| MeMYC 2.2-qF | CCCTGACCAGGGTGAGAATGAT | 实时荧光定量 RT-qPCR |
| MeMYC 2.2-qR | CCCTTTCGACACATGCTGAT | |
| AtActin1-qF | GGGCAAGTGATCACCATTGG | 转基因拟南芥基因表达分析 Gene expression analysis of transgenic Arabidopsis |
| AtActin1-qR | TGGAGCCAAAGCAGTGATCTC | |
| AtCBF1-qF | ATTATTGTCCGACGTTGGCCA | |
| AtCBF1-qR | GCGAAGTTGAGACATGCTGAT | |
| AtCBF2-qF | TGCCTGTCTCAATTTCGCTGA | |
| AtCBF2-qR | CGGCCATGTTATCCAACAAAC | |
| AtCBF3-qF | CAAGGATTTGGCTCGGAACAT | |
| AtCBF3-qR | TCCACCAACGTCTCCTCCATG | |
| AtCBF4-qF | AAGCTGCAATGGCGTTTCAGA | |
| AtCBF4-qR | CGTCACCCACTCCGTCAAAGT |
Table 1 Primers used in this study
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 引物用途 Primer usage |
|---|---|---|
| MeMYC 2.2-F | GCTCTAGAATGAATCTGT- GGACGGACGA | 基因克隆 Gene cloning |
| MeMYC 2.2-R | GTGGATCCGGAGTCACCA- ACTTTGGTTG | |
| MeActin-qF | TGATGAGTCTGGTCCATCCA | 内参基因 |
| MeActin-qR | CCTCCTACGACCCAATCTCA | Reference gene |
| MeMYC 2.2-qF | CCCTGACCAGGGTGAGAATGAT | 实时荧光定量 RT-qPCR |
| MeMYC 2.2-qR | CCCTTTCGACACATGCTGAT | |
| AtActin1-qF | GGGCAAGTGATCACCATTGG | 转基因拟南芥基因表达分析 Gene expression analysis of transgenic Arabidopsis |
| AtActin1-qR | TGGAGCCAAAGCAGTGATCTC | |
| AtCBF1-qF | ATTATTGTCCGACGTTGGCCA | |
| AtCBF1-qR | GCGAAGTTGAGACATGCTGAT | |
| AtCBF2-qF | TGCCTGTCTCAATTTCGCTGA | |
| AtCBF2-qR | CGGCCATGTTATCCAACAAAC | |
| AtCBF3-qF | CAAGGATTTGGCTCGGAACAT | |
| AtCBF3-qR | TCCACCAACGTCTCCTCCATG | |
| AtCBF4-qF | AAGCTGCAATGGCGTTTCAGA | |
| AtCBF4-qR | CGTCACCCACTCCGTCAAAGT |
| 分析内容 Item | MeMYC2.1 | MeMYC2.2 |
|---|---|---|
| 开放阅读框Open read frame(ORF) | 2 055 bp | 2 019 bp |
| 内含子 Intron | - | - |
| 蛋白质的分子量Molecular weight(Mw) | 75 043.89 | 74 015.17 |
| 等电点 Isoelectric point(pI) | 5.49 | 5.14 |
| Plant-mPLoc软件预测 Plant-mPLoc software prediction | 定位于细胞核 | 定位于细胞核 |
| SignalP | 非分泌蛋白 | 非分泌蛋白 |
| 保守功能结构域Conservative functional domains | bHLH-MYC保守结构域(N69-20),及HLH DNA结合域(N505-556) | bHLH-MYC保守结构域(N5-243),及HLH DNA结合域(N218-239) |
Table 2 Bioinformatics analysis of MeMYC2
| 分析内容 Item | MeMYC2.1 | MeMYC2.2 |
|---|---|---|
| 开放阅读框Open read frame(ORF) | 2 055 bp | 2 019 bp |
| 内含子 Intron | - | - |
| 蛋白质的分子量Molecular weight(Mw) | 75 043.89 | 74 015.17 |
| 等电点 Isoelectric point(pI) | 5.49 | 5.14 |
| Plant-mPLoc软件预测 Plant-mPLoc software prediction | 定位于细胞核 | 定位于细胞核 |
| SignalP | 非分泌蛋白 | 非分泌蛋白 |
| 保守功能结构域Conservative functional domains | bHLH-MYC保守结构域(N69-20),及HLH DNA结合域(N505-556) | bHLH-MYC保守结构域(N5-243),及HLH DNA结合域(N218-239) |
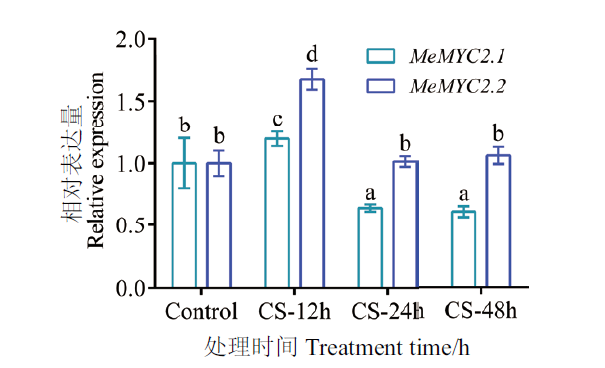
Fig. 1 Expression patterns of MeMYC2 under different cold stress(CS)time In vitro cassava seedling leaves were treated by low temperature at 10℃. New expanding leaves of stressed or unstressed seedlings were used in qPCR analysis. The MeActin1 was used as housekeep gene. Error bars are ± SD(n=3), different letters indicate significant differences with P≤0.05(ANOVA test), the same below
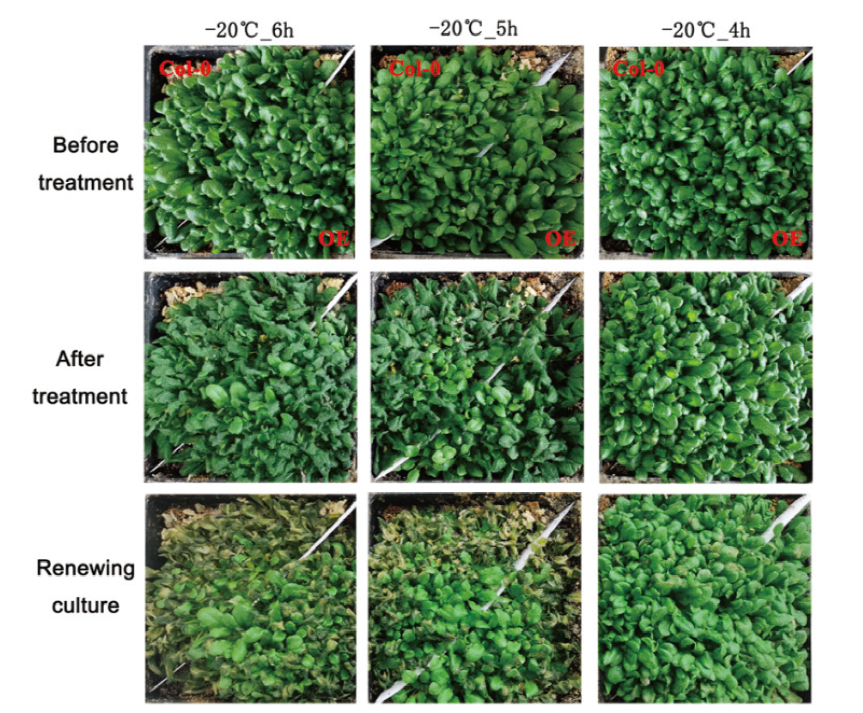
Fig. 4 Wild-type and transgenic plants demonstrating different freezing tolerance abilities Two-week-old seedlings were treated by cold at -20℃ for 4-6 h, and then these treated seedlings were recovered for 5 d under normal conditions
| 基因型 Genotype | 冷冻处理 Freezing treatment/h | 存活率 Survival rate/% |
|---|---|---|
| Col-0 | 4 | 100 |
| 5 | 74.2±1.5 | |
| 6 | 29.4±7.1 | |
| MYC2.2 OE | 4 | 100 |
| 5 | 92.2±2.3 | |
| 6 | 83.6±3.6 |
Table 3 Survival rates of wild-type and transgenic plants with cold treatment (n>30)
| 基因型 Genotype | 冷冻处理 Freezing treatment/h | 存活率 Survival rate/% |
|---|---|---|
| Col-0 | 4 | 100 |
| 5 | 74.2±1.5 | |
| 6 | 29.4±7.1 | |
| MYC2.2 OE | 4 | 100 |
| 5 | 92.2±2.3 | |
| 6 | 83.6±3.6 |
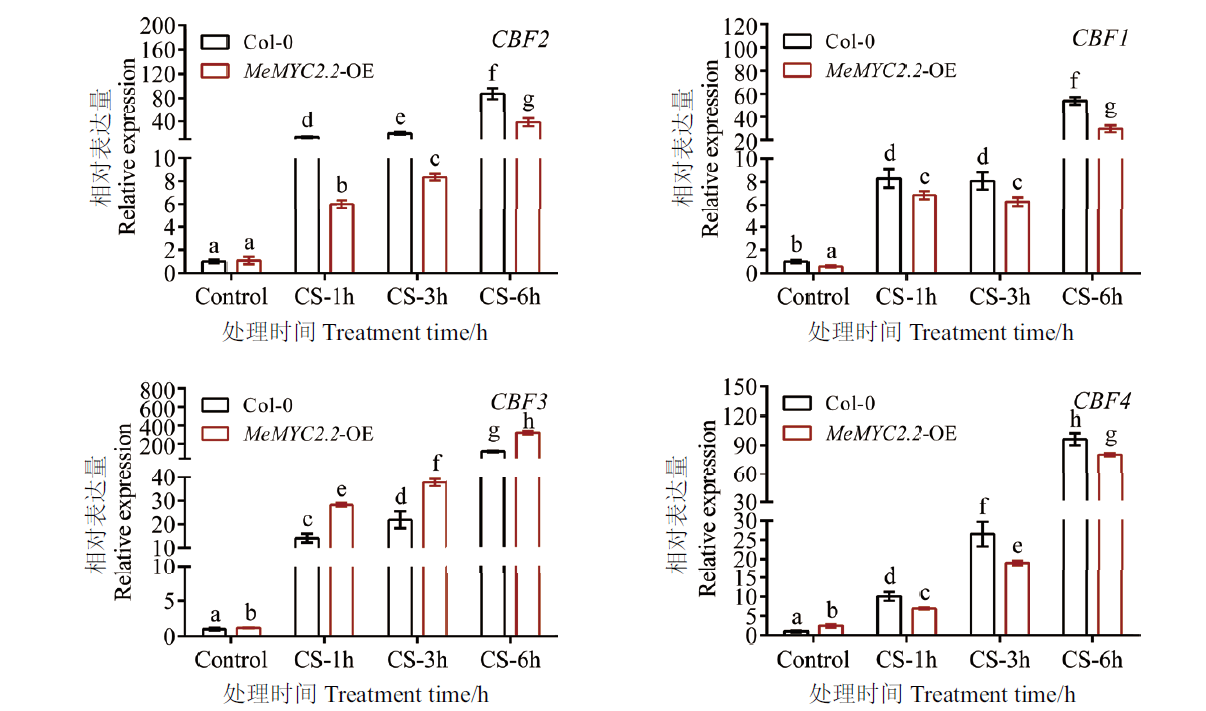
Fig. 5 Expression analysis of CBF genes in transgenic Arabidopsis under cold stress The relative expression of indicated CBF gene in Col-0 Arabidopsis under normal conditions was set to 1. Error bars are ± SD(n=3), and different letters indicate differences with P≤0.05(ANOVA test)
| [1] |
Shan ZY, Luo XL, Wu MY, et al. Genome-wide identification and expression of GRAS gene family members in cassava[J]. BMC Plant Biol, 2020, 20(1): 46.
doi: 10.1186/s12870-020-2242-8 pmid: 31996133 |
| [2] | 但忠. 广西木薯的抗寒性与抗寒育种的初步研究[D]. 海口: 海南大学, 2010. |
| Dan Z. The cold-resisdance of cassva and preliminary investigation for breeding of cassava[D]. Haikou: Hainan University, 2010. | |
| [3] | 黄洁, 李开绵, 叶剑秋, 等. 我国的木薯优势区域概述[J]. 广西农业科学, 2008, 39(1): 104-108. |
| Huang J, Li KM, Ye JQ, et al. A summary review of dominant regions of cassava growing in China[J]. Guangxi Agric Sci, 2008, 39(1): 104-108. | |
| [4] | 吴远航, 刘秦, 刘攀道, 等. 木薯苯丙氨酸解氨酶基因的克隆及其对低温胁迫的响应[J]. 热带作物学报, 2019, 40(3): 483-489. |
| Wu YH, Liu Q, Liu PD, et al. Cloning of cassava phenylalanine ammonia lyase genes and their responses to low temperature stress[J]. Chin J Trop Crops, 2019, 40(3): 483-489. | |
| [5] |
Xu J, Duan XG, Yang J, et al. Enhanced reactive oxygen species scavenging by overproduction of superoxide dismutase and catalase delays postharvest physiological deterioration of cassava storage roots[J]. Plant Physiol, 2013, 161(3): 1517-1528.
doi: 10.1104/pp.112.212803 pmid: 23344905 |
| [6] |
Xu J, Yang J, Duan XG, et al. Increased expression of native cytosolic Cu/Zn superoxide dismutase and ascorbate peroxidase improves tolerance to oxidative and chilling stresses in cassava(Manihot esculenta Crantz)[J]. BMC Plant Biol, 2014, 14: 208.
doi: 10.1186/s12870-014-0208-4 URL |
| [7] |
Cheng ZH, Lei N, Li SX, et al. The regulatory effects of MeTCP4 on cold stress tolerance in Arabidopsis thaliana: a transcriptome analysis[J]. Plant Physiol Biochem, 2019, 138: 9-16.
doi: 10.1016/j.plaphy.2019.02.015 URL |
| [8] | An D, Ma QX, Yan W, et al. Divergent regulation of CBF regulon on cold tolerance and plant phenotype in cassava overexpressing Arabidopsis CBF3 gene[J]. Front Plant Sci, 2016, 7: 1866. |
| [9] |
Ruan MB, Guo X, Wang B, et al. Genome-wide characterization and expression analysis enables identification of abiotic stress-responsive MYB transcription factors in cassava(Manihot esculenta)[J]. J Exp Bot, 2017, 68(13): 3657-3672.
doi: 10.1093/jxb/erx202 URL |
| [10] |
Omondi JO, Yermiyahu U, Rachmilevitch S, et al. Optimizing root yield of cassava under fertigation and the masked effect of atmospheric temperature[J]. J Sci Food Agric, 2020, 100(12): 4592-4600.
doi: 10.1002/jsfa.10519 URL |
| [11] | 李罡, 李文龙, 许雪梅, 等. MYC2转录因子参与植物发育调控的研究进展[J]. 植物生理学报, 2019, 55(2): 125-132. |
| Li G, Li WL, Xu XM, et al. Research progress of MYC2 transcription factors participating in plant development and regulation[J]. Plant Physiol J, 2019, 55(2): 125-132. | |
| [12] | 周文平, 王怀琴, 郭晓荣, 等. 丹参bHLH转录因子基因SmMYC2的克隆和表达分析[J]. 植物科学学报, 2016, 34(2): 246-254. |
| Zhou WP, Wang HQ, Guo XR, et al. Cloning and expression analysis of SmMYC2, a bHLH transcription factor gene from Salvia miltiorrhiza bunge[J]. Plant Sci J, 2016, 34(2): 246-254. | |
| [13] |
Hu YR, Jiang YJ, Han X, et al. Jasmonate regulates leaf senescence and tolerance to cold stress: crosstalk with other phytohormones[J]. J Exp Bot, 2017, 68(6): 1361-1369.
doi: 10.1093/jxb/erx004 pmid: 28201612 |
| [14] |
Hong GJ, Xue XY, Mao YB, et al. Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression[J]. Plant Cell, 2012, 24(6): 2635-2648.
doi: 10.1105/tpc.112.098749 URL |
| [15] |
Aleman F, Yazaki J, Lee M, et al. An ABA-increased interaction of the PYL6 ABA receptor with MYC2 Transcription Factor: a putative link of ABA and JA signaling[J]. Sci Rep, 2016, 6: 28941.
doi: 10.1038/srep28941 pmid: 27357749 |
| [16] |
Hiruma K, Nishiuchi T, Kato T, et al. Arabidopsis enhanced disease resistance 1 is required for pathogen-induced expression of plant defensins in nonhost resistance, and acts through interference of MYC2-mediated repressor function[J]. Plant J, 2011, 67(6): 980-992.
doi: 10.1111/j.1365-313X.2011.04651.x URL |
| [17] |
An JP, Wang XN, Yao JF, et al. Apple MdMYC2 reduces aluminum stress tolerance by directly regulating MdERF3 gene[J]. Plant Soil, 2017, 418(1/2): 255-266.
doi: 10.1007/s11104-017-3297-7 URL |
| [18] |
Zhao ML, Wang JN, Shan W, et al. Induction of jasmonate signalling regulators MaMYC2s and their physical interactions with MaICE1 in methyl jasmonate-induced chilling tolerance in banana fruit[J]. Plant Cell Environ, 2013, 36(1): 30-51.
doi: 10.1111/j.1365-3040.2012.02551.x URL |
| [19] | 于晓玲, 郭鑫, 李淑霞, 等. 逆境信号下木薯MeMYC2转录因子表达及功能分析[J]. 热带作物学报, 2021, 42(4): 927-935. |
| Yu XL, Guo X, Li SX, et al. Expression analysis of MeMYC2 transcription factor in cassava under stress signal[J]. Chin J Trop Crops, 2021, 42(4): 927-935. | |
| [20] |
An D, Yang J, Zhang P. Transcriptome profiling of low temperature-treated cassava apical shoots showed dynamic responses of tropical plant to cold stress[J]. BMC Genomics, 2012, 13: 64.
doi: 10.1186/1471-2164-13-64 pmid: 22321773 |
| [21] |
Chen QL, Li Z, Fan GZ, et al. Indications of stratospheric anomalies in the freezing rain and snow disaster in South China, 2008[J]. Sci China Earth Sci, 2011, 54(8): 1248-1256.
doi: 10.1007/s11430-011-4192-3 URL |
| [22] | Alves AAC. Cassava botany and physiology[M]// Hillocks RJ, Thresh JM, Bellotti AC. In Cassava: biology, production and utilization CAB International, 2002: 67-89. |
| [23] |
Meshi T, Iwabuchi M. Plant transcription factors[J]. Plant Cell Physiol, 1995, 36(8): 1405-1420.
pmid: 8589926 |
| [24] |
Jia YX, Ding YL, Shi YT, et al. The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis[J]. New Phytol, 2016, 212(2): 345-353.
doi: 10.1111/nph.14088 URL |
| [25] | Ritonga FN, Chen S. Physiological and molecular mechanism involved in cold stress tolerance in plants[J]. Plants(Basel), 2020, 9(5): 560. |
| [26] |
Zhao CZ, Zhang ZJ, Xie SJ, et al. Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis[J]. Plant Physiol, 2016, 171(4): 2744-2759.
doi: 10.1104/pp.16.00533 URL |
| [27] |
Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis -acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit[J]. Proc Natl Acad Sci USA, 1997, 94(3): 1035-1040.
doi: 10.1073/pnas.94.3.1035 pmid: 9023378 |
| [28] |
Gilmour SJ, Fowler SG, Thomashow MF. Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities[J]. Plant Mol Biol, 2004, 54(5): 767-781.
doi: 10.1023/B:PLAN.0000040902.06881.d4 pmid: 15356394 |
| [29] |
Hao JJ, Yang JL, Dong JL, et al. Characterization of BdCBF genes and genome-wide transcriptome profiling of BdCBF3-dependent and-independent cold stress responses in Brachypodium distachyon[J]. Plant Sci, 2017, 262: 52-61.
doi: 10.1016/j.plantsci.2017.06.001 URL |
| [30] |
Miura K, Jin JB, Lee J, et al. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis[J]. Plant Cell, 2007, 19(4): 1403-1414.
doi: 10.1105/tpc.106.048397 URL |
| [31] |
Fursova OV, Pogorelko GV, Tarasov VA. Identification of ICE2, a gene involved in cold acclimation which determines freezing tolerance in Arabidopsis thaliana[J]. Gene, 2009, 429(1/2): 98-103.
doi: 10.1016/j.gene.2008.10.016 URL |
| [32] |
Gilmour SJ, Zarka DG, Stockinger EJ, et al. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression[J]. Plant J, 1998, 16(4): 433-442.
doi: 10.1046/j.1365-313x.1998.00310.x pmid: 9881163 |
| [33] |
An D, Ma QX, Wang HX, et al. Cassava C-repeat binding factor 1 gene responds to low temperature and enhances cold tolerance when overexpressed in Arabidopsis and cassava[J]. Plant Mol Biol, 2017, 94(1/2): 109-124.
doi: 10.1007/s11103-017-0596-6 URL |
| [34] |
Yang YL, Liao WB, Yu XL, et al. Overexpression of MeDREB1D confers tolerance to both drought and cold stresses in transgenic Arabidopsis[J]. Acta Physiol Plant, 2016, 38(10): 1-11.
doi: 10.1007/s11738-015-2023-4 URL |
| [1] | CHEN Xiao, YU Ming-lan, WU Long-kun, ZHENG Xiao-ming, PANG Hong-bo. Research Progress in lncRNA and Their Responses to Low Temperature Stress in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 1-12. |
| [2] | XIAO Liang, WU Zheng-dan, LU Liu-ying, SHI Ping-li, SHANG Xiao-hong, CAO Sheng, ZENG Wen-dan, YAN Hua-bing. Research Progress of Important Traits Genes in Cassava [J]. Biotechnology Bulletin, 2023, 39(6): 31-48. |
| [3] | WANG Hai-long, LI Yu-qian, WANG Bo, XING Guo-fang, ZHANG Jie-wei. Isolation and Expression Analysis of SiMAPK3 in Setaria italica L. [J]. Biotechnology Bulletin, 2023, 39(3): 123-132. |
| [4] | YAO Xiao-wen, LIANG Xiao, CHEN Qing, WU Chun-ling, LIU Ying, LIU Xiao-qiang, SHUI Jun, QIAO Yang, MAO Yi-ming, CHEN Yin-hua, ZHANG Yin-dong. Study on the Expression Pattern of Genes in Lignin Biosynthesis Pathway of Cassava Resisting to Tetranychus urticae [J]. Biotechnology Bulletin, 2023, 39(2): 161-171. |
| [5] | ZHANG Xiao-yan, YANG Shu-hua, DING Yang-lin. Molecular Mechanism of Cold Signal Perception and Transduction in Plants [J]. Biotechnology Bulletin, 2023, 39(11): 28-35. |
| [6] | HAN Zhi-ling, CHEN Qing, LIANG Xiao, WU Chun-ling, LIU Ying, WU Mu-feng, XU Xue-lian. Influence on Expression of Jasmonic Acid Signaling Pathway Gene in Tetranychus urticae Fed on Mite-resistant and Mite-susceptible Cassava Cultivars [J]. Biotechnology Bulletin, 2022, 38(6): 211-220. |
| [7] | PAN Ying-jie, ZHANG Ying, WU Qi-man, LI Zheng-qing. A Review of WRKY Mediated Regulation of Sugar for Cold Acclimation in Horticultural Crops [J]. Biotechnology Bulletin, 2022, 38(3): 203-212. |
| [8] | ZOU Liang-ping, GUO Xin, QI Deng-feng, ZHAI Min, LI Zhuang, ZHAO Ping-juan, PENG Ming, NIU Xing-kui. Anthocyanin Accumulation and Its Gene Expression Induced by Low Nitrogen Stress in Cassava Seedlings [J]. Biotechnology Bulletin, 2022, 38(2): 75-82. |
| [9] | LIU Li-li, ZHU Hua, YAN Yan-chun, WANG Xiao-wen, ZHANG Rong, ZHU Jian-ya. Research Progress of Cold Tolerance Mechanism and Functional Genes in Fish [J]. Biotechnology Bulletin, 2018, 34(8): 50-57. |
| [10] | WANG Ya-ru ,LIANG Xiao ,WU Chun-ling ,CHEN Qing ,ZHAO Hui-ping. Activity Variations of Protective Enzymes in Paracoccus marginatus After Fed Different Cassava Cultivars [J]. Biotechnology Bulletin, 2018, 34(6): 115-119. |
| [11] | DING Ze-hong, FU Li-li, YAN Yan, TIE Wei-wei, HU Wei. Evolution Analysis of P5CR and Expression Analysis of P5CR Genes in Cassava [J]. Biotechnology Bulletin, 2018, 34(3): 105-112. |
| [12] | XIAO Yu-jie, LI Ze-ming, YI Peng-fei, HU Ri-sheng, ZHANG Xian-wen, ZHU Lie-shu. Research Progress on Response Mechanism of Transcription Factors Involved in Plant Cold Stress [J]. Biotechnology Bulletin, 2018, 34(12): 1-9. |
| [13] | TANG Shi-yun, YANG Li-tao, LI Yang-rui. Comparative Analysis on Transcriptome Among Different Sugarcane Cultivars Under Low Temperature Stress [J]. Biotechnology Bulletin, 2018, 34(12): 116-124. |
| [14] | LIANG Xiao, LU Fu-ping, LU Hui, WU Chun-ling, CAO Xian-hong, CHEN Qing. Preliminary Study on the Function of Polyphenol Oxidase in Cassava Resistance to Mite [J]. Biotechnology Bulletin, 2017, 33(4): 143-148. |
| [15] | SONG Yan-chao, An Fei-fei, Xue Jing-jing, Qin Yu-ling, Li Kai-mian, CHEN Song-bi. Proteomic Analysis on Tuberous Roots of Cassava Cultivar ZM-Seaside and Mosaic-leaf Mutation [J]. Biotechnology Bulletin, 2017, 33(3): 78-85. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||