








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (10): 292-303.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0281
Previous Articles Next Articles
ZHANG Ao-jie1( ), LI Qing-yun1,2(
), LI Qing-yun1,2( ), SONG Wen-hong1, YAN Shao-hui1, TANG Ai-xing1,2, LIU You-yan1,2
), SONG Wen-hong1, YAN Shao-hui1, TANG Ai-xing1,2, LIU You-yan1,2
Received:2023-03-27
Online:2023-10-26
Published:2023-11-28
Contact:
LI Qing-yun
E-mail:zhangaojieao@163.com;qyli@gxu.edu.cn
ZHANG Ao-jie, LI Qing-yun, SONG Wen-hong, YAN Shao-hui, TANG Ai-xing, LIU You-yan. Whole Genome Sequencing Analysis of a Phenol-degrading Strain Alcaligenes faecalis JF101[J]. Biotechnology Bulletin, 2023, 39(10): 292-303.
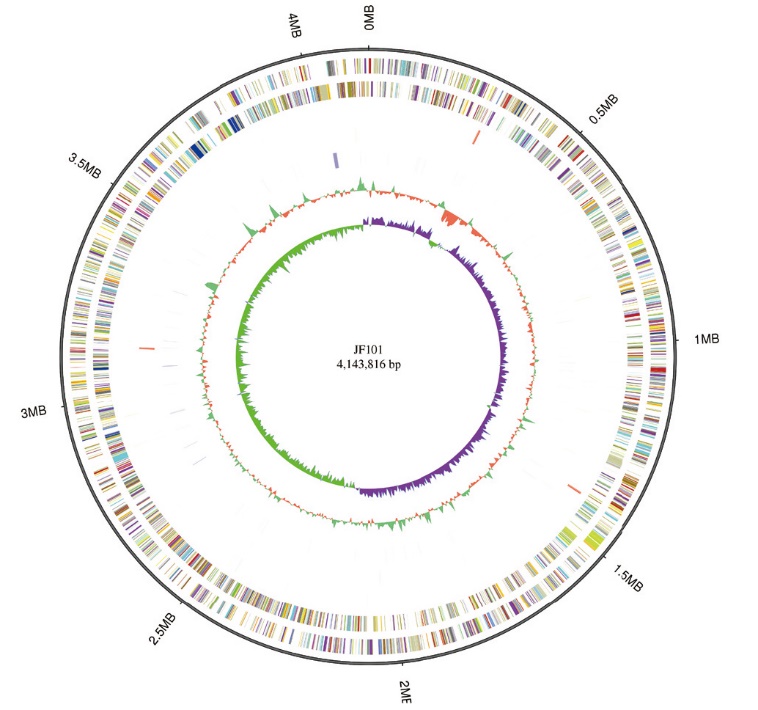
Fig. 1 Circular representation of Alcaligenes faecalis JF101 genome From the outside to the inside, the circle chart shows the genome size, forward strand gene, reverse strand gene, forward strand ncRNA, reverse strand ncRNA, repeat sequence, GC share, and GC offset
| Protein | Amino acid | Proposed function | Sequence similarrty | Identity similarity/% | Accession No |
|---|---|---|---|---|---|
| GL-1751 | 396 | Muconate cycloisomerase | catB, Alcaligenes faecalis strain ZD02 | 97.0 | ALO39907.1 |
| GL-1752 | 311 | Catechol 1,2-dioxygenase | catA, Alcaligenes faecalis strain ZD02 | 99.7 | ALO39908.1 |
| GL-1755 | 267 | 3-oxoadipate enol-lactonase | pcaD, Alcaligenes aquatilis strain QD168 | 98.5 | AYN20737.1 |
| GL-1756 | 91 | Muconolactone D-isomerase | catC, Alcaligenes faecalis strain ZD02 | 100 | ALO39912.1 |
| GL-1849 | 334 | Nif-specific regulatory protein | MOPR, Sediminispirochaeta smaragdinae DSM 11293 | 47.9 | ADK82111.1 |
| GL-1850 | 82 | Phenol hydroxylase P0 protein | dmpK, Alcaligenes faecalis strain JQ135 | 97.6 | ASR89481.1 |
| GL-1851 | 332 | Phenol hydroxylase P1 protein | dmpL, Alcaligenes faecalis strain ZD02 | 99.1 | ALO39961.1 |
| GL-1852 | 90 | Phenol hydroxylase P2 protein | dmpM, Alcaligenes faecalis strain ZD02 | 100 | ALO39962.1 |
| GL-1853 | 498 | Phenol hydroxylase P3 protein | dmpN, Psychrobacter sp. P2G3 | 86.7 | AMN49709.1 |
| GL-1854 | 120 | Phenol hydroxylase P4 protein | dmpO, Alcaligenes faecalis strain ZD02 | 98.3 | ALO39964.1 |
| GL-1855 | 353 | Phenol hydroxylase P5 protein | dmpP, Alcaligenes faecalis strain ZD02 | 98.9 | ALO39965.1 |
Table 1 Homology comparison of phenol degradation-related genes of A. faecalis JF101 and homologous genes in other bacteria
| Protein | Amino acid | Proposed function | Sequence similarrty | Identity similarity/% | Accession No |
|---|---|---|---|---|---|
| GL-1751 | 396 | Muconate cycloisomerase | catB, Alcaligenes faecalis strain ZD02 | 97.0 | ALO39907.1 |
| GL-1752 | 311 | Catechol 1,2-dioxygenase | catA, Alcaligenes faecalis strain ZD02 | 99.7 | ALO39908.1 |
| GL-1755 | 267 | 3-oxoadipate enol-lactonase | pcaD, Alcaligenes aquatilis strain QD168 | 98.5 | AYN20737.1 |
| GL-1756 | 91 | Muconolactone D-isomerase | catC, Alcaligenes faecalis strain ZD02 | 100 | ALO39912.1 |
| GL-1849 | 334 | Nif-specific regulatory protein | MOPR, Sediminispirochaeta smaragdinae DSM 11293 | 47.9 | ADK82111.1 |
| GL-1850 | 82 | Phenol hydroxylase P0 protein | dmpK, Alcaligenes faecalis strain JQ135 | 97.6 | ASR89481.1 |
| GL-1851 | 332 | Phenol hydroxylase P1 protein | dmpL, Alcaligenes faecalis strain ZD02 | 99.1 | ALO39961.1 |
| GL-1852 | 90 | Phenol hydroxylase P2 protein | dmpM, Alcaligenes faecalis strain ZD02 | 100 | ALO39962.1 |
| GL-1853 | 498 | Phenol hydroxylase P3 protein | dmpN, Psychrobacter sp. P2G3 | 86.7 | AMN49709.1 |
| GL-1854 | 120 | Phenol hydroxylase P4 protein | dmpO, Alcaligenes faecalis strain ZD02 | 98.3 | ALO39964.1 |
| GL-1855 | 353 | Phenol hydroxylase P5 protein | dmpP, Alcaligenes faecalis strain ZD02 | 98.9 | ALO39965.1 |
| 菌株 Strain | 基因组大小 Genome size/Mb | 编码蛋白 Protein coding | rRNA数量 rRNA genes | tRNA数量 tRNA genes | GC含量 GC/% | 质粒数 Number of plasmids |
|---|---|---|---|---|---|---|
| MUB14 | 4.35 | 3973 | 9 | 58 | 56.74 | 2(0.04 Mb, 0.06 Mb) |
| P156 | 4.04 | 3616 | 9 | 57 | 56.7 | 0 |
| ZD02 | 4.23 | 3752 | 9 | 57 | 56.81 | 1(0.01 Mb) |
| c16 | 4.30 | 3847 | 9 | 57 | 56.3 | 0 |
| ASM96730v2 | 4.23 | 3770 | 9 | 57 | 56.8 | 1(0.01 Mb) |
| JF101 | 4.14 | 3804 | 9 | 55 | 57.44 | 0 |
Table 2 Basic characteristics of the genome of A. faecalis
| 菌株 Strain | 基因组大小 Genome size/Mb | 编码蛋白 Protein coding | rRNA数量 rRNA genes | tRNA数量 tRNA genes | GC含量 GC/% | 质粒数 Number of plasmids |
|---|---|---|---|---|---|---|
| MUB14 | 4.35 | 3973 | 9 | 58 | 56.74 | 2(0.04 Mb, 0.06 Mb) |
| P156 | 4.04 | 3616 | 9 | 57 | 56.7 | 0 |
| ZD02 | 4.23 | 3752 | 9 | 57 | 56.81 | 1(0.01 Mb) |
| c16 | 4.30 | 3847 | 9 | 57 | 56.3 | 0 |
| ASM96730v2 | 4.23 | 3770 | 9 | 57 | 56.8 | 1(0.01 Mb) |
| JF101 | 4.14 | 3804 | 9 | 55 | 57.44 | 0 |
| 相容性溶质转运相关基因 Compatible solutes transport genes | 基因名称 Name | 基因位置 Locus tag | 基因长度 Length/bp | 基因描述 Description |
|---|---|---|---|---|
| K+ transport system | Trk system Kdp system | GL-1953、GL-1954 GL-1574、GL-1575、GL-1576 | 455、217 568、237、687 | Trk系统钾摄取蛋白 K+转运ATP酶A、B、C链 |
| Proline and betaine transport | ProP system | GL-0834、GL-1501、GL-2239、GL-2295、GL-3438、GL-3579 | 452、436、565、423、425、510 | MFS转运体,MHS家族,脯氨酸/甜菜碱转运体 |
| Choline/glycine/proline betaine transport genes | betT、betS | GL-2836 | 706 | 胆碱/甘氨酸/脯氨酸甜菜碱转运蛋白 |
Table 3 Salt-tolerant genes related to compatible solute transport in strain JF101
| 相容性溶质转运相关基因 Compatible solutes transport genes | 基因名称 Name | 基因位置 Locus tag | 基因长度 Length/bp | 基因描述 Description |
|---|---|---|---|---|
| K+ transport system | Trk system Kdp system | GL-1953、GL-1954 GL-1574、GL-1575、GL-1576 | 455、217 568、237、687 | Trk系统钾摄取蛋白 K+转运ATP酶A、B、C链 |
| Proline and betaine transport | ProP system | GL-0834、GL-1501、GL-2239、GL-2295、GL-3438、GL-3579 | 452、436、565、423、425、510 | MFS转运体,MHS家族,脯氨酸/甜菜碱转运体 |
| Choline/glycine/proline betaine transport genes | betT、betS | GL-2836 | 706 | 胆碱/甘氨酸/脯氨酸甜菜碱转运蛋白 |
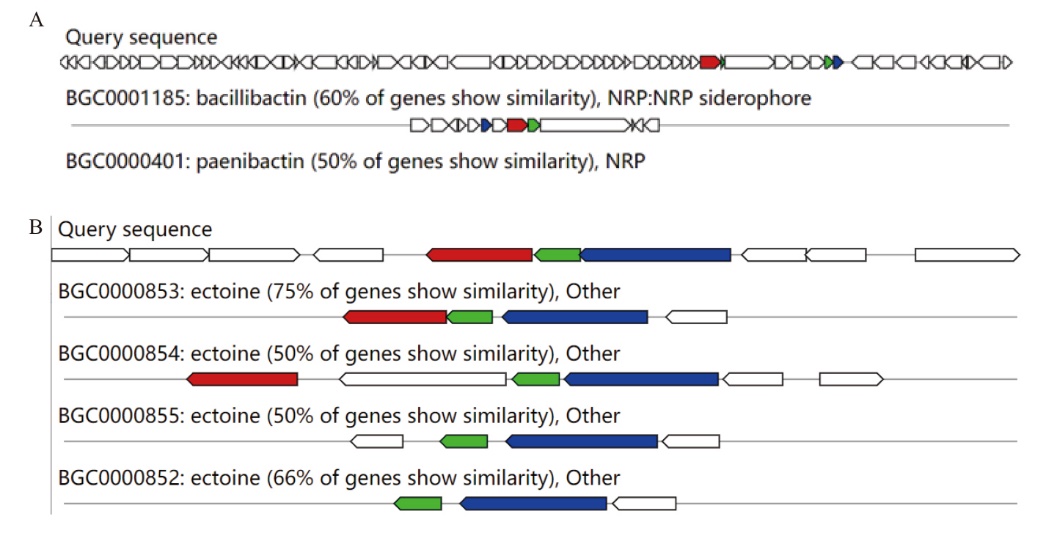
Fig. 7 Sequence comparison of the JF101 secondary metabolite synthesis gene cluster in A. faecalis JF101 A: Bacteriocin gene cluste. B: Terpene synthesis gene cluste
| [1] |
Pazarlioğlu NK, Telefoncu A. Biodegradation of phenol by Pseudomonas putida immobilized on activated pumice particles[J]. Process Biochem, 2005, 40(5): 1807-1814.
doi: 10.1016/j.procbio.2004.06.043 URL |
| [2] |
Amor L, Eiroa M, Kennes C, et al. Phenol biodegradation and its effect on the nitrification process[J]. Water Res, 2005, 39(13): 2915-2920.
pmid: 15998531 |
| [3] |
Li XY, Cui YH, Feng YJ, et al. Reaction pathways and mechanisms of the electrochemical degradation of phenol on different electrodes[J]. Water Res, 2005, 39(10): 1972-1981.
doi: 10.1016/j.watres.2005.02.021 URL |
| [4] |
Liu L, Si L, Yang JH, et al. Biodegradation and process optimization of phenol and formaldehyde by Aspergillus nomius SGFA1[J]. Int Biodeterior Biodegrad, 2023, 182: 105630.
doi: 10.1016/j.ibiod.2023.105630 URL |
| [5] |
Saravanan P, Pakshirajan K, Saha P. Growth kinetics of an indigenous mixed microbial consortium during phenol degradation in a batch reactor[J]. Bioresour Technol, 2008, 99(1): 205-209.
doi: 10.1016/j.biortech.2006.11.045 URL |
| [6] |
Wang CC, Lee CM, Lu CJ, et al. Biodegradation of 2, 4, 6-trichlorophenol in the presence of primary substrate by immobilized pure culture bacteria[J]. Chemosphere, 2000, 41(12): 1873-1879.
pmid: 11061309 |
| [7] |
Prieto MB, Hidalgo A, Rodríguez-Fernández C, et al. Biodegradation of phenol in synthetic and industrial wastewater by Rhodococcus erythropolis UPV-1 immobilized in an air-stirred reactor with clarifier[J]. Appl Microbiol Biotechnol, 2002, 58(6): 853-859.
pmid: 12021809 |
| [8] |
Santos VL, Linardi VR. Biodegradation of phenol by a filamentous fungi isolated from industrial effluents—identification and degradation potential[J]. Process Biochem, 2004, 39(8): 1001-1006.
doi: 10.1016/S0032-9592(03)00201-2 URL |
| [9] |
Arutchelvan V, Kanakasabai V, Elangovan R, et al. Kinetics of high strength phenol degradation using Bacillus brevis[J]. J Hazard Mater, 2006, 129(1/2/3): 216-222.
doi: 10.1016/j.jhazmat.2005.08.040 URL |
| [10] |
Jiang Y, Wen JP, Bai J, et al. Biodegradation of phenol at high initial concentration by Alcaligenes faecalis[J]. J Hazard Mater, 2007, 147(1/2): 672-676.
doi: 10.1016/j.jhazmat.2007.05.031 URL |
| [11] |
Rehfuss M, Urban J. Alcaligenes faecalis subsp. phenolicus subsp. nov. a phenol-degrading, denitrifying bacterium isolated from a graywater bioprocessor[J]. Syst Appl Microbiol, 2005, 28(5): 421-429.
pmid: 16094869 |
| [12] |
Kumar A, Bhunia B, Dasgupta D, et al. Optimization of culture condition for growth and phenol degradation by Alcaligenes faecalisJF339228 using Taguchi Methodology[J]. Desalin Water Treat, 2013, 51(16/17/18): 3153-3163.
doi: 10.1080/19443994.2012.749021 URL |
| [13] |
Deveryshetty J, Phale PS. Biodegradation of phenanthrene by Alcaligenes sp. strain PPH: partial purification and characterization of 1-hydroxy-2-naphthoic acid hydroxylase[J]. FEMS Microbiol Lett, 2010, 311(1): 93-101.
doi: 10.1111/j.1574-6968.2010.02079.x pmid: 20727010 |
| [14] |
Bharali P, Das S, Konwar BK, et al. Crude biosurfactant from thermophilic Alcaligenes faecalis: feasibility in petro-spill bioremediation[J]. Int Biodeterior Biodegrad, 2011, 65(5): 682-690.
doi: 10.1016/j.ibiod.2011.04.001 URL |
| [15] |
John RC, Essien JP, Akpan SB, et al. Polycyclic aromatic hydrocarbon-degrading bacteria from aviation fuel spill site at Ibeno, Nigeria[J]. Bull Environ Contam Toxicol, 2012, 88(6): 1014-1019.
doi: 10.1007/s00128-012-0598-7 URL |
| [16] |
Shah PD, Dave SR, Rao MS. Enzymatic degradation of textile dye Reactive Orange 13 by newly isolated bacterial strain Alcaligenes faecalis PMS-1[J]. Int Biodeterior Biodegrad, 2012, 69: 41-50.
doi: 10.1016/j.ibiod.2012.01.002 URL |
| [17] |
Siripattanakul S, Wirojanagud W, McEvoy J, et al. Atrazine degradation by stable mixed cultures enriched from agricultural soil and their characterization[J]. J Appl Microbiol, 2009, 106(3): 986-992.
doi: 10.1111/j.1365-2672.2008.04075.x pmid: 19191954 |
| [18] | Kong LF, Zhu SY, Zhu LS, et al. Biodegradation of organochlorine pesticide endosulfan by bacterial strain Alcaligenes faecalis JBW4[J]. J Environ Sci(China), 2013, 25(11): 2257-2264. |
| [19] |
Pandeeti EVP, Siddavattam D. Purification and characterization of catechol 1, 2-dioxygenase from Acinetobacter sp. DS002 and cloning, sequencing of partial catA gene[J]. Indian J Microbiol, 2011, 51(3): 312-318.
doi: 10.1007/s12088-011-0123-4 URL |
| [20] |
Tuan NN, Hsieh HC, Lin YW, et al. Analysis of bacterial degradation pathways for long-chain alkylphenols involving phenol hydroxylase, alkylphenol monooxygenase and catechol dioxygenase genes[J]. Bioresour Technol, 2011, 102(5): 4232-4240.
doi: 10.1016/j.biortech.2010.12.067 URL |
| [21] | 陈健峰, 徐天虹, 梁谏婷, 等. 2, 4-二氯苯酚羟化酶在大肠杆菌中的表达及靛蓝的生物合成[J]. 药物生物技术, 2015, 22(6): 476-479. |
| Chen JF, Xu TH, Liang JT, et al. Cloning and expression of 2, 4-dichlorophenol hydroxylase and indigo biosynthesis in Escherichia coli[J]. Pharm Biotechnol, 2015, 22(6): 476-479. | |
| [22] |
Zhou WG, Guo WB, Zhou HB, et al. Phenol degradation by Sulfobacillus acidophilus TPY via the meta-pathway[J]. Microbiol Res, 2016, 190: 37-45.
doi: 10.1016/j.micres.2016.05.005 URL |
| [23] |
Trivedi VD, Jangir PK, Sharma R, et al. Erratum: insights into functional and evolutionary analysis of carbaryl metabolic pathway from Pseudomonas sp. strain C5pp[J]. Sci Rep, 2017, 7: 40899.
doi: 10.1038/srep40899 pmid: 28358360 |
| [24] |
Silva CC, Hayden H, Sawbridge T, et al. Phylogenetic and functional diversity of metagenomic libraries of phenol degrading sludge from petroleum refinery wastewater treatment system[J]. AMB Express, 2012, 2(1): 18.
doi: 10.1186/2191-0855-2-18 pmid: 22452812 |
| [25] |
Shingler V, Powlowski J, Marklund U. Nucleotide sequence and functional analysis of the complete phenol/3, 4-dimethylphenol catabolic pathway of Pseudomonas sp. strain CF600[J]. J Bacteriol, 1992, 174(3): 711-724.
pmid: 1732207 |
| [26] |
Teramoto M, Ohnishi K, Harayama S, et al. An AraC/XylS family member at a high level in a hierarchy of regulators for phenol-metabolizing enzymes in Comamonas testosteroni R5[J]. J Bacteriol, 2002, 184(14): 3941-3946.
doi: 10.1128/JB.184.14.3941-3946.2002 pmid: 12081966 |
| [27] |
Powlowski J, Shingler V. In vitro analysis of polypeptide requirements of multicomponent phenol hydroxylase from Pseudomonas sp. strain CF600[J]. J Bacteriol, 1990, 172(12): 6834-6840.
pmid: 2254259 |
| [28] | 苏佳岐, 张红兵. 苯酚羟化酶基因的研究进展[J]. 产业与科技论坛, 2017, 16(14): 48-50. |
| Su JQ, Zhang HB. Research progress of phenol hydroxylase gene[J]. Ind Sci Tribune, 2017, 16(14): 48-50. | |
| [29] |
Lagesen K, Hallin P, Rødland EA, et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes[J]. Nucleic Acids Res, 2007, 35(9): 3100-3108.
doi: 10.1093/nar/gkm160 pmid: 17452365 |
| [30] |
Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence[J]. Nucleic Acids Res, 1997, 25(5): 955-964.
doi: 10.1093/nar/25.5.955 pmid: 9023104 |
| [31] |
Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches[J]. Bioinformatics, 2013, 29(22): 2933-2935.
doi: 10.1093/bioinformatics/btt509 pmid: 24008419 |
| [32] |
Nawrocki EP, Burge SW, Bateman A, et al. Rfam 12.0: updates to the RNA families database[J]. Nucleic Acids Res, 2015, 43(Database issue): D130-D137.
doi: 10.1093/nar/gku1063 URL |
| [33] |
Benson G. Tandem repeats finder: a program to analyze DNA sequences[J]. Nucleic Acids Res, 1999, 27(2): 573-580.
doi: 10.1093/nar/27.2.573 pmid: 9862982 |
| [34] |
Nandi T, Ong C, Singh AP, et al. A genomic survey of positive selection in Burkholderia pseudomallei provides insights into the evolution of accidental virulence[J]. PLoS Pathog, 2010, 6(4): e1000845.
doi: 10.1371/journal.ppat.1000845 URL |
| [35] |
Konuk HB, Ergüden B. Phenolic-OH group is crucial for the antifungal activity of terpenoids via disruption of cell membrane integrity[J]. Folia Microbiol, 2020, 65(4): 775-783.
doi: 10.1007/s12223-020-00787-4 |
| [36] |
Park KH, Kim S, Lee SJ, et al. Tetrameric architecture of an active phenol-bound form of the AAA+ transcriptional regulator DmpR[J]. Nat Commun, 2020, 11(1): 2728.
doi: 10.1038/s41467-020-16562-5 |
| [37] |
Suvorova IA, Gelfand MS. Comparative genomic analysis of the regulation of aromatic metabolism in betaproteobacteria[J]. Front Microbiol, 2019, 10: 642.
doi: 10.3389/fmicb.2019.00642 pmid: 30984152 |
| [38] |
Yu HY, Peng ZX, Zhan YH, et al. Novel regulator MphX represses activation of phenol hydroxylase genes caused by a XylR/DmpR-type regulator MphR in Acinetobacter calcoaceticus[J]. PLoS One, 2011, 6(3): e17350.
doi: 10.1371/journal.pone.0017350 URL |
| [39] | 彭子欣, 于海英, 战嵛华, 等. 苯酚羟化酶的功能与调控研究进展[J]. 中国农业科技导报, 2010, 12(5): 36-41. |
| Peng ZX, Yu HY, Zhan YH, et al. Advances in function and regulation of phenol hydroxylase[J]. J Agric Sci Technol, 2010, 12(5): 36-41. | |
| [40] |
Arai H, Akahira S, Ohishi T, et al. Adaptation of Comamonas testosteroni TA441 to utilization of phenol by spontaneous mutation of the gene for a trans-acting factor[J]. Mol Microbiol, 1999, 33(6): 1132-1140.
pmid: 10510228 |
| [41] |
Qu YY, Shi SN, Zhou H, et al. Characterization of a novel phenol hydroxylase in indoles biotransformation from a strain Arthrobacter sp. W1[J]. PLoS One, 2012, 7(9): e44313.
doi: 10.1371/journal.pone.0044313 URL |
| [42] |
Sela I, Wolf YI, Koonin EV. Assessment of assumptions underlying models of prokaryotic pangenome evolution[J]. BMC Biology, 2021, 19(1): 27.
doi: 10.1186/s12915-021-00960-2 pmid: 33563283 |
| [43] | 卢源达, 陈玲, 杜云龙, 等. 不同禾本科作物中ZmWRKY79同源基因的鉴定与分析[J]. 西南农业学报, 2023, 36(3): 522-531. |
| Lu YD, Chen L, Du YL, et al. Identification and analysis of ZmWRKY79 homologous genes in different gramineal crops[J]. Southwest China J Agric Sci, 2023, 36(3): 522-531. | |
| [44] |
Majewski P, Majewska P, Gutowska A, et al. Molecular characterisation of clinical pandrug-resistant Alcaligenes faecalis strain MUB14[J]. Int J Antimicrob Agents, 2020, 55(6): 105939.
doi: 10.1016/j.ijantimicag.2020.105939 URL |
| [45] |
Hu CH, Zhao SX, Li KR, et al. Microbial degradation of nicotinamide by a strain Alcaligenes sp. P156[J]. Sci Rep, 2019, 9(1): 3647.
doi: 10.1038/s41598-019-40199-0 |
| [46] |
Ju SY, Zheng JS, Lin J, et al. The complete genome sequence of Alcaligenes faecalis ZD02, a novel potential bionematocide[J]. J Biotechnol, 2016, 218: 73-74.
doi: 10.1016/j.jbiotec.2015.12.001 URL |
| [47] |
Kong LF, Zhu SY, Zhu LS, et al. Colonization of Alcaligenes faecalis strain JBW4 in natural soils and its detoxification of endosulfan[J]. Appl Microbiol Biotechnol, 2014, 98(3): 1407-1416.
doi: 10.1007/s00253-013-5033-4 URL |
| [48] |
Liu YX, Wang Y, Li Y, et al. Nitrogen removal characteristics of heterotrophic nitrification-aerobic denitrification by Alcaligenes faecalis C16[J]. Chin J Chem Eng, 2015, 23(5): 827-834.
doi: 10.1016/j.cjche.2014.04.005 URL |
| [49] |
Finks SS, Martiny JBH. Plasmid-encoded traits vary across environments[J]. mBio, 2023, 14(1): e0319122.
doi: 10.1128/mbio.03191-22 URL |
| [50] |
Alva VA, Peyton BM. Phenol and catechol biodegradation by the haloalkaliphile Halomonas campisalis: influence of pH and salinity[J]. Environ Sci Technol, 2003, 37(19): 4397-4402.
doi: 10.1021/es0341844 URL |
| [51] |
Li H, Meng FP, Duan WY, et al. Biodegradation of phenol in saline or hypersaline environments by bacteria: a review[J]. Ecotoxicol Environ Saf, 2019, 184: 109658.
doi: 10.1016/j.ecoenv.2019.109658 URL |
| [52] |
Tanudjaja E, Hoshi N, Yamamoto K, et al. Two Trk/Ktr/HKT-type potassium transporters, TrkG and TrkH, perform distinct functions in Escherichia coli K-12[J]. J Biol Chem, 2023, 299(2): 102846.
doi: 10.1016/j.jbc.2022.102846 URL |
| [53] |
Malek AA, Chen CL, Wargo MJ, et al. Roles of three transporters, CbcXWV, BetT1, and BetT3, in Pseudomonas aeruginosa choline uptake for catabolism[J]. J Bacteriol, 2011, 193(12): 3033-3041.
doi: 10.1128/JB.00160-11 pmid: 21478341 |
| [54] | Souii A, Gorrab A, Ouertani R, et al. Sustainable bioethanol production from enzymatically hydrolyzed second-generation Posidonia oceanica waste using stable Microbacterium metallidurans carbohydrate-active enzymes as biocatalysts[J]. Biomass Convers Biorefin, 2022: 1-20. |
| [55] |
Mei RW, Zhou M, Xu LN, et al. Characterization of a pH-tolerant strain Cobetia sp. SASS1 and its phenol degradation performance under salinity condition[J]. Front Microbiol, 2019, 10: 2034.
doi: 10.3389/fmicb.2019.02034 URL |
| [56] |
Gong Y, Ding P, Xu MJ, et al. Biodegradation of phenol by a halotolerant versatile yeast Candida tropicalis SDP-1 in wastewater and soil under high salinity conditions[J]. J Environ Manage, 2021, 289: 112525.
doi: 10.1016/j.jenvman.2021.112525 URL |
| [57] |
Zhang SS, An ZJ, Su XM, et al. Phenol degradation at high salinity by a resuscitated strain Pseudomonas sp. SAS26: kinetics and pathway[J]. J Environ Chem Eng, 2023, 11(4): 110182.
doi: 10.1016/j.jece.2023.110182 URL |
| [1] | WANG Teng-hui, GE Wen-dong, LUO Ya-fang, FAN Zhen-yu, WANG Yu-shu. Gene Mapping of Kale White Leaves Based on Whole Genome Re-sequencing of Extreme Mixed Pool(BSA) [J]. Biotechnology Bulletin, 2023, 39(9): 176-182. |
| [2] | FANG Lan, LI Yan-yan, JIANG Jian-wei, CHENG Sheng, SUN Zheng-xiang, ZHOU Yi. Isolation, Identification and Growth-promoting Characteristics of Endohyphal Bacterium 7-2H from Endophytic Fungi of Spiranthes sinensis [J]. Biotechnology Bulletin, 2023, 39(8): 272-282. |
| [3] | GUO Shao-hua, MAO Hui-li, LIU Zheng-quan, FU Mei-yuan, ZHAO Ping-yuan, MA Wen-bo, LI Xu-dong, GUAN Jian-yi. Whole Genome Sequencing and Comparative Genome Analysis of a Fish-derived Pathogenic Aeromonas Hydrophila Strain XDMG [J]. Biotechnology Bulletin, 2023, 39(8): 291-306. |
| [4] | ZHANG Zhi-xia, LI Tian-pei, ZENG Hong, ZHU Xi-xian, YANG Tian-xiong, MA Si-nan, HUANG Lei. Genome Sequencing and Bioinformatics Analysis of Gelidibacter sp. PG-2 [J]. Biotechnology Bulletin, 2023, 39(3): 290-300. |
| [5] | HE Meng-ying, LIU Wen-bin, LIN Zhen-ming, LI Er-tong, WANG Jie, JIN Xiao-bao. Whole Genome Sequencing and Analysis of an Anti Gram-positive Bacterium Gordonia WA4-43 [J]. Biotechnology Bulletin, 2023, 39(2): 232-242. |
| [6] | WANG Shuai, LV Hong-rui, ZHANG Hao, WU Zhan-wen, XIAO Cui-hong, SUN Dong-mei. Whole-Genome Sequencing Identification of Phosphate-solubilizing Bacteria PSB-R and Analysis of Its Phosphate-solubilizing Properties [J]. Biotechnology Bulletin, 2023, 39(1): 274-283. |
| [7] | WEN Chang, LIU Chen, LU Shi-yun, XU Zhong-bing, AI Chao-fan, LIAO Han-peng, ZHOU Shun-gui. Biological Characteristics and Genome Analysis of a Novel Multidrug-resistant Shigella flexneri Phage [J]. Biotechnology Bulletin, 2022, 38(9): 127-135. |
| [8] | LI Ji-hong, JING Yu-ling, MA Gui-zhen, GUO Rong-jun, LI Shi-dong. Genome Construction of Achromobacter 77 and Its Characteristics on Chemotaxis and Antibiotic Resistance [J]. Biotechnology Bulletin, 2022, 38(9): 136-146. |
| [9] | ZHANG Ze-ying, FAN Qing-feng, DENG Yun-feng, WEI Ting-zhou, ZHOU Zheng-fu, ZHOU Jian, WANG Jin, JIANG Shi-jie. Whole Genome Sequencing and Comparative Genomic Analysis of a High-yield Lipase-producing Strain WCO-9 [J]. Biotechnology Bulletin, 2022, 38(10): 216-225. |
| [10] | CHEN Ti-qiang, XU Xiao-lan, SHI Lin-chun, ZHONG Li-Yi. Sequencing and Analysis of the Whole Genome of Zizhi Cultivar ‘Wuzhi No.2’(Ganoderma sp. strain Zizhi S2) [J]. Biotechnology Bulletin, 2021, 37(11): 42-56. |
| [11] | GUO He-bao, WANG Xing, HE Shan-wen, ZHANG Xiao-xia. Phenotypic Characteristics Combined with Genomic Analysis to Identify Different Colony Morphology Bacillus velezensis ACCC 19742 [J]. Biotechnology Bulletin, 2020, 36(2): 142-148. |
| [12] | XIANG Sha, LIU Ming-xue, ZHANG Ge-ge, LUO Lang, WEI Hong-fu, DONG Fa-qin. Screening of Photoelectron-response Microbes as well as Their Growth and Metabolism [J]. Biotechnology Bulletin, 2017, 33(4): 205-213. |
| [13] | Wang Xingwen,Wang Jiaqi,Zhao Shengguo,Li Fadi,Bu Dengpan. The Application of Uncultured Methods in the Study of Ruminal Methanogen Population [J]. Biotechnology Bulletin, 2014, 0(6): 67-74. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||