








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (12): 128-135.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0539
Previous Articles Next Articles
HOU Lin-hui( ), ZHENG Yun, RONG Er-hua, WU Yu-xiang(
), ZHENG Yun, RONG Er-hua, WU Yu-xiang( )
)
Received:2023-06-08
Online:2023-12-26
Published:2024-01-11
Contact:
WU Yu-xiang
E-mail:wan19980816hlh@163.com;yuxiangwu2009@hotmail.com
HOU Lin-hui, ZHENG Yun, RONG Er-hua, WU Yu-xiang. Character Identification and Genetic Analysis of Progeny from Distant Hybrid Between Gossypium hirsutum and G. thurberi[J]. Biotechnology Bulletin, 2023, 39(12): 128-135.
| 类名 Classific name | 染色体组 Genome | 染色体数Number of chromosomes | 来源 Source |
|---|---|---|---|
| 陆地棉 G. hirsutum | AD1 | 2n=4X=52 | 中国农科院棉花研究所 Cotton Research Institute, CAAS |
| 瑟伯氏棉 G. thurberi | D1 | 2n=2X=26 | 国家种质三亚野生棉圃 Sanya National Germplasm Wild Cotton Garden |
Table 1 Basic information of research materials
| 类名 Classific name | 染色体组 Genome | 染色体数Number of chromosomes | 来源 Source |
|---|---|---|---|
| 陆地棉 G. hirsutum | AD1 | 2n=4X=52 | 中国农科院棉花研究所 Cotton Research Institute, CAAS |
| 瑟伯氏棉 G. thurberi | D1 | 2n=2X=26 | 国家种质三亚野生棉圃 Sanya National Germplasm Wild Cotton Garden |
| 正向引物Forward primer | 引物序列Primer sequence(5'-3') | 反向引物Reverse primer | 引物序列Primer sequence(5'-3') |
|---|---|---|---|
| BNL4108-F | TCCACCATTCCCGTAAATGT | BNL4108-R | TGGCCAAGTCATTAGGCTTT |
| BNL4053-F | TGAAGGCTTTGAAGCAAACA | BNL4053-R | AAGCAAGCACCAAGTTAGCC |
| NAU2026-F | GAATCTCGAAAACCCCATCT | NAU2026-R | ATTTGGAAGCGAAGTACCAG |
| NAU1355-F | ATCTGTTTACGCCACTCTCC | NAU1355-R | CCAGCCTTTGACATTTTTCT |
| NAU1169-F | GGGTAGTAGCTTTTATGATAGGG | NAU1169-R | CCATTCCTTCCCCTAATTCT |
| NAU1157-F | GAGTTTGGTTCTGGGTTGAG | NAU1157-R | GATCCTTTTCATCTCCTCCA |
| NAU1052-F | CGCAGATAAAGGATGGATTT | NAU1052-R | AGAGCTGGAGGACATAACAAA |
| NAU1042-F | CATGCAAATCCATGCTAGAG | NAU1042-R | GGTTTCTTTGGTGGTGAAAC |
| NAU1164-F | CCAACGCTAATTCTACCTCCT | NAU1164-R | GCGGGTAATTGTAGTACATGC |
| NBRI_GI1015-F | GAGTTTGGCTCCGAGTTGAG | NBRI_GI1015-R | GCGAGTGAACTCACCGATAA |
| NBRI_GM7-F | GTGATACCTGGAAAGATGTGA | NBRI_GM7-R | TCCTGATGCTGTTTTAGTCTT |
| NBRI_Gpd_21-F | GCAATTGTGGAAGAGAAGTAA | NBRI_Gpd_21-R | GGACCTCTGAGTCCATTTATT |
| NBRI_Gpd_85-F | CTCAAAAACCCCTTTAGTCTC | NBRI_Gpd_85-R | GAGTGAAAACCCGCAAAG |
Table 2 SSR primers sequence information
| 正向引物Forward primer | 引物序列Primer sequence(5'-3') | 反向引物Reverse primer | 引物序列Primer sequence(5'-3') |
|---|---|---|---|
| BNL4108-F | TCCACCATTCCCGTAAATGT | BNL4108-R | TGGCCAAGTCATTAGGCTTT |
| BNL4053-F | TGAAGGCTTTGAAGCAAACA | BNL4053-R | AAGCAAGCACCAAGTTAGCC |
| NAU2026-F | GAATCTCGAAAACCCCATCT | NAU2026-R | ATTTGGAAGCGAAGTACCAG |
| NAU1355-F | ATCTGTTTACGCCACTCTCC | NAU1355-R | CCAGCCTTTGACATTTTTCT |
| NAU1169-F | GGGTAGTAGCTTTTATGATAGGG | NAU1169-R | CCATTCCTTCCCCTAATTCT |
| NAU1157-F | GAGTTTGGTTCTGGGTTGAG | NAU1157-R | GATCCTTTTCATCTCCTCCA |
| NAU1052-F | CGCAGATAAAGGATGGATTT | NAU1052-R | AGAGCTGGAGGACATAACAAA |
| NAU1042-F | CATGCAAATCCATGCTAGAG | NAU1042-R | GGTTTCTTTGGTGGTGAAAC |
| NAU1164-F | CCAACGCTAATTCTACCTCCT | NAU1164-R | GCGGGTAATTGTAGTACATGC |
| NBRI_GI1015-F | GAGTTTGGCTCCGAGTTGAG | NBRI_GI1015-R | GCGAGTGAACTCACCGATAA |
| NBRI_GM7-F | GTGATACCTGGAAAGATGTGA | NBRI_GM7-R | TCCTGATGCTGTTTTAGTCTT |
| NBRI_Gpd_21-F | GCAATTGTGGAAGAGAAGTAA | NBRI_Gpd_21-R | GGACCTCTGAGTCCATTTATT |
| NBRI_Gpd_85-F | CTCAAAAACCCCTTTAGTCTC | NBRI_Gpd_85-R | GAGTGAAAACCCGCAAAG |

Fig. 1 Comparison of flower anatomy and stamens of different generations P1: G. hirsutum; P2: G. thurber; F1: hybrid; M1: allohexaploid. The same below

Fig. 4 A comparison of meiosis behaviors in different generations(Bar=20 μm) A-E are F1(A: Monad. B: Dyad and two Micronucleus. C: Triad. D: Tetrad.E: Polyad). F-J are M1(F: Monad. G: Triad. H: Abnormal Tetrad. I: Normal Tetrad.J: Polyad
| 倍性 Ploidy | 单分体 Monad | 二分体 Dyad | 三分体 Triad | 四分体 Tetrad | 多分体 Polyad | 总数 Total |
|---|---|---|---|---|---|---|
| F1(3X) | 73(12.17) | 25(4.17) | 38(6.33) | 146(24.33) | 318(53.00) | 600 |
| M1(6X) | 63(10.50) | 15(2.50) | 17(2.83) | 493(82.17) | 12(2.00) | 600 |
Table 3 Comparison of meiosis in different generations
| 倍性 Ploidy | 单分体 Monad | 二分体 Dyad | 三分体 Triad | 四分体 Tetrad | 多分体 Polyad | 总数 Total |
|---|---|---|---|---|---|---|
| F1(3X) | 73(12.17) | 25(4.17) | 38(6.33) | 146(24.33) | 318(53.00) | 600 |
| M1(6X) | 63(10.50) | 15(2.50) | 17(2.83) | 493(82.17) | 12(2.00) | 600 |
| 材料(倍性) Material(Ploidy) | 气孔密度 Stomatal density | 气孔比 Porosity ratio | 叶绿体数Number of chloroplasts |
|---|---|---|---|
| F1(3X) | 36.35±1.55a | 1.28±0.17a | 16.45±2.54a |
| M1(6X) | 26.45±1.89b | 1.40±0.12b | 26.31±2.30b |
Table 4 Comparison of stomatas in different generations
| 材料(倍性) Material(Ploidy) | 气孔密度 Stomatal density | 气孔比 Porosity ratio | 叶绿体数Number of chloroplasts |
|---|---|---|---|
| F1(3X) | 36.35±1.55a | 1.28±0.17a | 16.45±2.54a |
| M1(6X) | 26.45±1.89b | 1.40±0.12b | 26.31±2.30b |
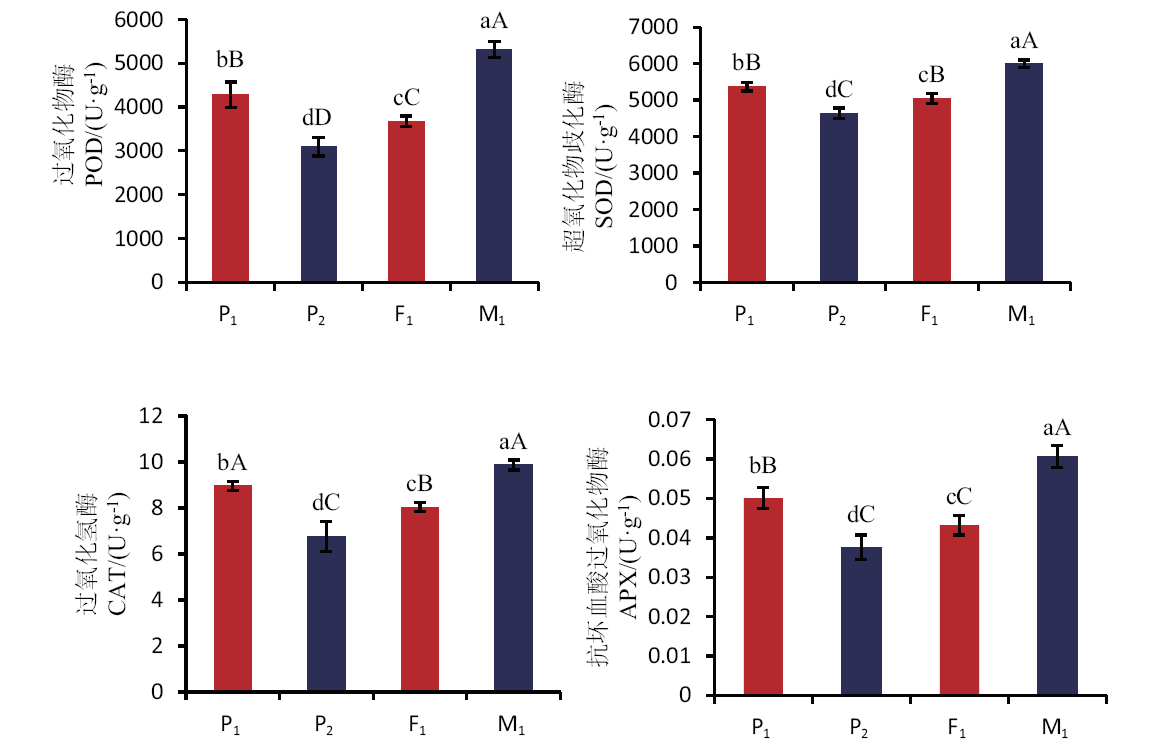
Fig. 6 Comparison of enzyme activities in different generations Lowercase letters indicate a significant difference at the 0.05 level, the capital letters indicate a significant difference at the 0.01 level. The same below
| 材料Material | 3X | 6X | ||||||
|---|---|---|---|---|---|---|---|---|
| 母本 Female parent | 父本 Male parent | 重组片段Recombination | 母本 Female parent | 父本 Male parent | 重组片段Recombination | |||
| SSR条带SSR bands | 34 | 21 | 4 | 34 | 21 | 6 | ||
| 遗传比例Genetic ratio/% | 57.62 | 35.59 | 6.79 | 55.73 | 34.42 | 9.85 | ||
Table 5 Analysis of SSR polymorphism in different generations
| 材料Material | 3X | 6X | ||||||
|---|---|---|---|---|---|---|---|---|
| 母本 Female parent | 父本 Male parent | 重组片段Recombination | 母本 Female parent | 父本 Male parent | 重组片段Recombination | |||
| SSR条带SSR bands | 34 | 21 | 4 | 34 | 21 | 6 | ||
| 遗传比例Genetic ratio/% | 57.62 | 35.59 | 6.79 | 55.73 | 34.42 | 9.85 | ||
| [1] | 王坤波. 野生棉的收集与保存[J]. 棉花学报, 2007, 19(5): 354-361. |
| Wang KB. Introduction and conservation of wild cotton in China[J]. Cotton Sci, 2007, 19(5): 354-361. | |
| [2] | 董琪. D基因组野生棉响应大丽轮枝菌侵染的转录组分析[D]. 保定: 河北农业大学, 2019. |
| Dong Q. Transcriptome analysis of D genome wild cotton in response to Verticillium dahlia infection[D]. Baoding: Hebei Agricultural University, 2019. | |
| [3] |
Sattler MC, Carvalho CR, Clarindo WR. The polyploidy and its key role in plant breeding[J]. Planta, 2016, 243(2): 281-296.
doi: 10.1007/s00425-015-2450-x pmid: 26715561 |
| [4] |
Mammadov J, Buyyarapu R, Guttikonda SK, et al. Wild relatives of maize, rice, cotton, and soybean: Treasure troves for tolerance to biotic and abiotic stresses[J]. Front Plant Sci, 2018, 9: 886.
doi: 10.3389/fpls.2018.00886 pmid: 30002665 |
| [5] |
Zhao FA, Fang WP, et al. Proteomic identification of differentially expressed proteins in Gossypium thurberi inoculated with cotton Verticillium dahliae[J]. Plant Sci, 2012, 185/186: 176-184.
doi: 10.1016/j.plantsci.2011.10.007 URL |
| [6] |
牛永章, 李炳林, 张原根, 等. 陆地棉×瑟伯氏棉杂种后代主要经济性状的遗传分析[J]. 华北农学报, 1989(1): 41-47.
doi: 10.3321/j.issn:1000-7091.1989.01.008 |
| Niu YZ, Li BL, Zhang YG, et al. Genetic analysis of the main economic characters of the hybrid progeny of upland cotton×Thurber cotton[J]. Acta Agri Boreali Sin, 1989(1): 41-47. | |
| [7] | 潘家驹, 闵留芳, 刘康, 等. 陆地棉芽黄基因应用于杂种棉的研究[J]. 南京农业大学学报, 1998, 21(3): 7-14. |
| Pan JJ, Min LF, Liu K, et al. The exploitation of virescent genes on upland hybrid cotton[J]. J Nanjing Agric Univ, 1998, 21(3): 7-14. | |
| [8] | 梁理民, 王增信, 等. 棉花种间杂交新品系及新种质的育成[J]. 西北农林科技大学学报: 自然科学版, 2003, 31(5):9-12. |
| Liang LM, Wang ZX, et al. Selection for new interspecific hybridization strains and germplasms of cotton[J]. J Northwest Sci Tech Univ Agric For Nat Sci Ed, 2003, 31(5): 9-12. | |
| [9] | 李爱国, 赵丽芬, 赵国忠. 远缘杂交棉花品种石远321的选育研究[J]. 安徽农业科学, 2006, 34(9): 1812-1813, 1815. |
| Li AG, Zhao LF, Zhao GZ. Study on selection and breeding of distant hybridization cotton variety Shiyuan 321[J]. J Anhui Agric Sci, 2006, 34(9): 1812-1813, 1815. | |
| [10] |
Cai X, Magwanga RO, Xu Y, et al. Comparative transcriptome, physiological and biochemical analyses reveal response mechanism mediated by CBF4 and ICE2 in enhancing cold stress tolerance in Gossypium thurberi[J]. AoB Plants, 2019, 11(6): plz045.
doi: 10.1093/aobpla/plz045 URL |
| [11] |
Gan Y, Chen D, Liu F, et al. Individual chromosome assignment and chromosomal collinearity in Gossypium thurberi, G. trilobum and D subgenome of G. barbadense revealed by BAC-FISH[J]. Genes Genet Syst, 2011, 86(3): 165-174.
doi: 10.1266/ggs.86.165 URL |
| [12] | Nakano Y, Asada K. Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts[J]. Plant Cell Physiol, 1981, 22(5): 867-880. |
| [13] | 李合生. 植物生理生化实验原理和技术[M]. 北京: 高等教育出版社, 2000. |
| Li HS. Principles and techniques of plant physiological biochemical experiment[M]. Beijing: Higher Education Press, 2000. | |
| [14] | 申状状, 杨娜, 等. 亚洲棉与瑟伯氏棉种间远缘杂种的合成及其性状鉴定[J]. 分子植物育种, 2017, 15(6): 2291-2297. |
| Shen ZZ, Yang N, et al. Synthesis and identification of interspecific hybrid between Gossypium arboreum L. and G. thurberi T.[J]. Mol Plant Breed, 2017, 15(6): 2291-2297. | |
| [15] | 宋平, 王志安, 王志宁. 棉属中染色体倍性与叶片气孔性状的关系[J]. 浙江农业大学学报, 1989(1): 39-44. |
| Song P, Wang ZA, Wang ZN. Relationship between chromosome ploidyand stomatal characters of leaf in Gossypium[J]. J Zhejiang Agric Univ, 1989, 15(1): 39-44. | |
| [16] | 胡绍安, 崔荣霞, 等. 陆地棉野生种系的利用研究[J]. 棉花学报, 1994, 6(S1): 15-18. |
| Hu SA, Cui RX, et al. Utilization and studies on wild races of G. hirsutum L.[J]. Acta Gossypii Sin, 1994, 6(S1): 15-18. | |
| [17] |
Cheng F, Wu J, Cai X, et al. Gene retention, fractionation and subgenome differences in polyploid plants[J]. Nat Plants, 2018, 4(5):258-268.
doi: 10.1038/s41477-018-0136-7 pmid: 29725103 |
| [18] |
申状状, 李昱樱, 等. 陆地棉和野生斯特提棉种间异源六倍体的合成与性状鉴定[J]. 作物学报, 2019, 45(4): 628-634.
doi: 10.3724/SP.J.1006.2019.84086 |
| Shen ZZ, Li YY, et al. Allohexaploid synthesis and its characteristic identification between cotton species Gossypium hirsutum and G. sturtianum[J]. Crop J, 2019, 45(4): 628-634. | |
| [19] | 王坤波, 刘旭. 棉属多倍化研究进展[J]. 中国农业科技导报, 2013, 15(2): 20-27. |
| Wang KB, Liu X. Study progress of Gossypium polyploidization[J]. J Agric Sci Technol, 2013, 15(2): 20-27. | |
| [20] |
Yin X, Zhan R, He Y, et al. Morphological description of a novel synthetic allotetraploid(A1A1G3G3)of Gossypium herbaceum L. and G. nelsonii Fryx suitable for disease-resistant breeding applications[J]. PLoS One, 2020, 15(12): e0242620.
doi: 10.1371/journal.pone.0242620 URL |
| [21] |
Chen D, Wu Y, Zhang X, et al. Analysis of[G. capitisviridis ×(G. hirsutum × G. australe)2]trispeci-fic hybrid and selected characteristics[J]. PLoS One, 2015, 10(6): e0127023.
doi: 10.1371/journal.pone.0127023 URL |
| [22] |
Meng S, Xu Z, Xu P, et al. A complete set of monosomic alien addition lines developed from Gossypium anomalum in a Gossypium hirsutum background: genotypic and phenotypic characterization[J]. Breed Sci, 2020, 70(4): 494-501.
doi: 10.1270/jsbbs.19146 URL |
| [23] | Zheng J, Kong X, et al. Comparative transcriptome analysis between a novel allohexaploid cotton progeny CMS line LD6A and its maintainer line LD6B[J]. Int J Mol Sci, 2019, 20(24): E6127. |
| [24] |
Masoomi-Aladizgeh F, Kamath KS, Haynes PA, et al. Genome survey sequencing of wild cotton(Gossypium robinsonii)reveals insights into proteomic responses of pollen to extreme heat[J]. Plant Cell Environ, 2022, 45(4): 1242-1256.
doi: 10.1111/pce.v45.4 URL |
| [25] |
Wei Y, Xu Y, Lu P, et al. Salt stress responsiveness of a wild cotton species(Gossypium klotzschianum)based on transcriptomic analysis[J]. PLoS One, 2017, 12(5): e0178313.
doi: 10.1371/journal.pone.0178313 URL |
| [1] | YIN Ming-hua, YU Huan-yuan, XIAO Xin-yi, WANG Yu-ting. Chloroplast Genomic Characterization and Phylogenetic Analysis of Colocasia esculenta L. Schoot var. cormosus cv. ‘Hongyayu’ from Jiangxi Yanshan [J]. Biotechnology Bulletin, 2023, 39(6): 233-247. |
| [2] | WANG Yi-fan, HOU Lin-hui, CHANG Yong-chun, YANG Ya-jie, CHEN Tian, ZHAO Zhu-yue, RONG Er-hua, WU Yu-xiang. Synthesis and Character Identification of Allohexaploid Between Gossypium hirsutum and G. gossypioides [J]. Biotechnology Bulletin, 2023, 39(5): 168-176. |
| [3] | AN Miao, WANG Tong-tong, FU Yi-ting, XIA Jun-jun, PENG Suo-tang, DUAN Yong-hong. Genetic Diversity Analysis and Molecular Identity Card Construction by SSR Markers of 52 Solanum tuberosum L. Varieties(Lines) [J]. Biotechnology Bulletin, 2023, 39(12): 136-147. |
| [4] | LU Yu-sheng, PENG Cheng, CHANG Xiao-xiao, QIU Ji-shui, CHEN Zhe, CHEN Hui-qiong. Genetic Diversity Analysis and Molecular Identity Establishment of Clausena lansium Germplasm Resources from Guangdong Province Based on SSR Markers [J]. Biotechnology Bulletin, 2023, 39(12): 187-199. |
| [5] | YANG Ya-jie, LI Yu-ying, SHEN Zhuang-zhuang, CHEN Tian, RONG Er-hua, WU Yu-xiang. Selection and Character Identification for Autopolyploid Progenies of Gossypium herbaceum [J]. Biotechnology Bulletin, 2022, 38(5): 64-73. |
| [6] | FU Yong-yao, YI De-yan, YANG Xian-mao, CAI Li, LIANG Yu-hua, LEI Mei-yan, YANG Li-ping. Analysis of Morphological Characteristics and Genetic Variation in a New Germplasm Lilium lancifolium JD-h-15 [J]. Biotechnology Bulletin, 2022, 38(11): 140-150. |
| [7] | ZHAO Zhu-yue, SHEN Zhuang-zhuang, WANG Yi-fan, YANG Ya-jie, RONG Er-hua, WU Yu-xiang. Character Identification and Genetic Analysis of Distant Hybrid Between Gossypium hirsutum and Gossypium sturtianum [J]. Biotechnology Bulletin, 2021, 37(5): 19-27. |
| [8] | GUAN Jun-jiao, YANG Xiao-hong, ZHANG Peng, HUANG Qing-mei, ZHANG Jian-hua. Establishment of a Method Identifying the Varieties of Hulless Barley Based on Fluorescence Detection Technology [J]. Biotechnology Bulletin, 2021, 37(11): 293-302. |
| [9] | WANG Yan-li, YANG Yi-ming, FAN Shu-tian, ZHAO Ying, XU Pei-lei, LU Wen-peng, LI Chang-yu. Genetic Diversity Analysis of 73 Vitis amurensis and Its Hybrids Offsprings Based on SSR Molecular Markers [J]. Biotechnology Bulletin, 2021, 37(1): 189-197. |
| [10] | WANG Yi-fan, ZHENG Yun, RONG Er-hua, WU Yu-xiang. Synthesis and Identification of Distant Hybrids Between Gossypium arboreum and Gossypium gossypioides [J]. Biotechnology Bulletin, 2020, 36(8): 1-7. |
| [11] | WáNG Yu, DENG Meng-sheng, XU Chi, YU Li-ping, XIE Liáng-shuái, LI Li-qin, WáNG Xi-yáo. Cloning ánd Expression ánálysis of StSSR2 Gene in Solánum tuberosum [J]. Biotechnology Bulletin, 2020, 36(4): 61-69. |
| [12] | LI Yong-hui, YU Xiáng-li, Má Hui-ping, GáO Kái, LIU Ming-xue. ánálysis on Genetic Diversity of ISSR from Different Páeoniá suffruticosá Cultivárs [J]. Biotechnology Bulletin, 2020, 36(4): 78-83. |
| [13] | ZHAO Yang-yang, GUO Yu-xiao, ZHANG Ling-yun. Transcriptome Sequencing and Analysis of Xanthoceras sorbifolia Bunge Fruit [J]. Biotechnology Bulletin, 2019, 35(6): 24-31. |
| [14] | LIU Pei-pei, LUO Guang-ming, CHAI Hua-wen, ZHANG Jun-yi, ZHU Zi-hao, WANG De-yun. Application of SSR Marker Technique in the Identification of Traditional Chinese Medicine [J]. Biotechnology Bulletin, 2019, 35(2): 198-203. |
| [15] | WANG Ping, WANG Chun-yu, ZHANG Li-xia, CONG Ling, ZHU Zhen-xing, LU Xiao-chun. Development of SSR Molecular Markers with Sorghum Polymorphism Using Re-sequencing [J]. Biotechnology Bulletin, 2019, 35(11): 217-223. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||