








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (2): 243-253.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0611
Previous Articles Next Articles
LI Wang-ning1( ), ZHANG Hao-jie1, LI Ya-nan1, LIANG Meng-jing1, JI Chun-li1, Zhang Chun-hui1, LI Run-zhi1, CUI Yu-lin2, QIN Song2, CUI Hong-li1(
), ZHANG Hao-jie1, LI Ya-nan1, LIANG Meng-jing1, JI Chun-li1, Zhang Chun-hui1, LI Run-zhi1, CUI Yu-lin2, QIN Song2, CUI Hong-li1( )
)
Received:2022-05-18
Online:2023-02-26
Published:2023-03-07
LI Wang-ning, ZHANG Hao-jie, LI Ya-nan, LIANG Meng-jing, JI Chun-li, Zhang Chun-hui, LI Run-zhi, CUI Yu-lin, QIN Song, CUI Hong-li. Phenotypic Characterization of Blue Photoreceptor Plant Type Cryptochrome CRY Mutant in Chlamydomonas reinhardtii[J]. Biotechnology Bulletin, 2023, 39(2): 243-253.
| 引物名称Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|
| Actin-F | AGAAGGACTCGTACGTTGGC |
| Actin-R | CCAGAGTCCAGCACGATACC |
| cry-F | AGCACCCAATGTGATACCCG |
| cry-R | CATACAGCATGTCGCCGTTG |
| Cassette-C1 | ATACTGCATGTAATGGCCAGG |
| Cassette-C2 | GCGGCAGAATAGTCGCGTAT |
Table 1 Primers information
| 引物名称Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|
| Actin-F | AGAAGGACTCGTACGTTGGC |
| Actin-R | CCAGAGTCCAGCACGATACC |
| cry-F | AGCACCCAATGTGATACCCG |
| cry-R | CATACAGCATGTCGCCGTTG |
| Cassette-C1 | ATACTGCATGTAATGGCCAGG |
| Cassette-C2 | GCGGCAGAATAGTCGCGTAT |
| 试剂Reagent | 用量Dosage/μL |
|---|---|
| TB Green Premix Ex Taq II(TliRNaseH Plus)(2×) | 5 |
| 正向引物 | 0.25 |
| 反向引物 | 0.25 |
| DNA模板 | 0.5 |
| 双蒸水 | 4 |
| 反应总体系 | 10 |
Table 2 PCR reaction system
| 试剂Reagent | 用量Dosage/μL |
|---|---|
| TB Green Premix Ex Taq II(TliRNaseH Plus)(2×) | 5 |
| 正向引物 | 0.25 |
| 反向引物 | 0.25 |
| DNA模板 | 0.5 |
| 双蒸水 | 4 |
| 反应总体系 | 10 |
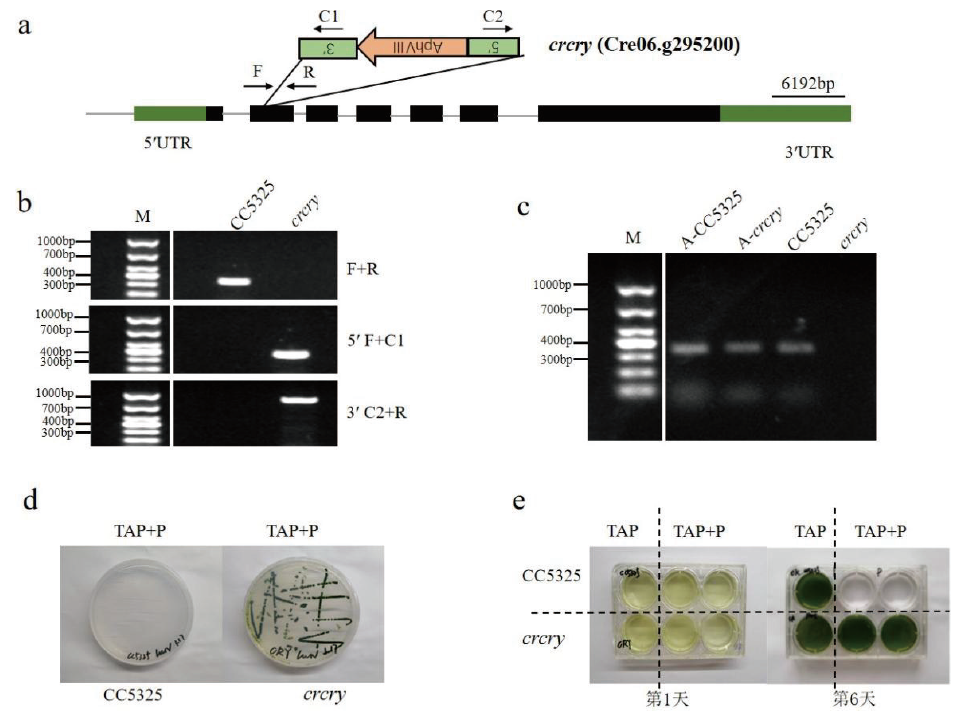
Fig.1 Identification of C. Reinhardtii crcry mutant a: Schematic map of the inserted cassette. b: PCR confirmation of insertion site DNA level. c: PCR validation of RNA levels at insertion sites. d: Plate screening of mutant strains. e: Liquid screening of mutant strains. P: Paromomycin

Fig. 3 Effects of blue light on the single cell pigments of C. reinhardtii CC5325 and crcry a: Chlorophyll a content in a single cell. b: Chlorophyll b content in a single cell. c: Content of chlorophyll c in a single cell. d: Content of chlorophyll d in a single cell. e: Chlorophyll e content in a single cell. f: Chlorophyll f content in a single cell
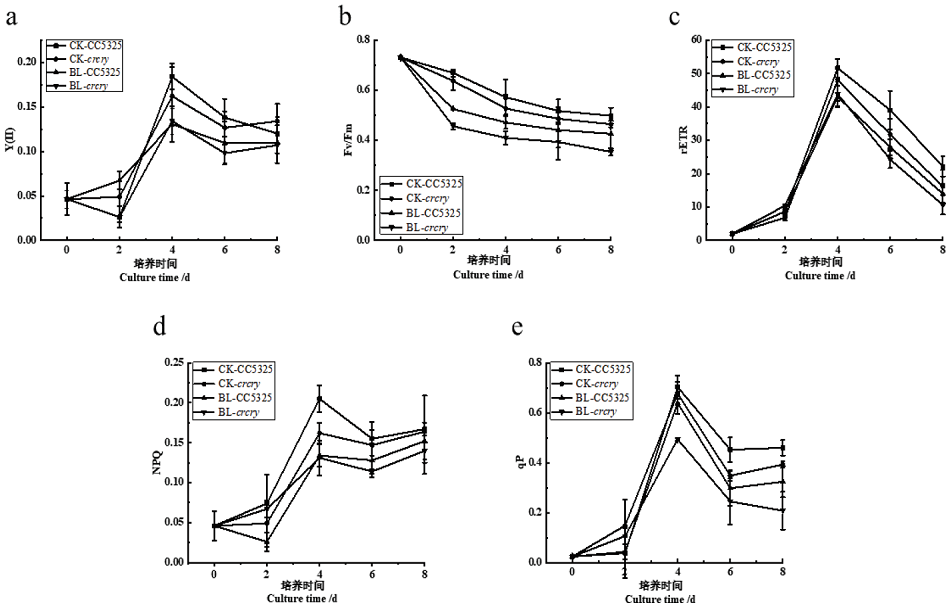
Fig. 4 Effects of blue light on the photosynthesis of C. Reinhardtii CC5325 and crcry a: Actual photochemical efficiency of photosystem II. b: Maximum photochemical efficiency of Fv/Fm. c: Relative electron transfer rate. d: Non-photochemical quenching coefficient. e: Photochemical quenching coefficient
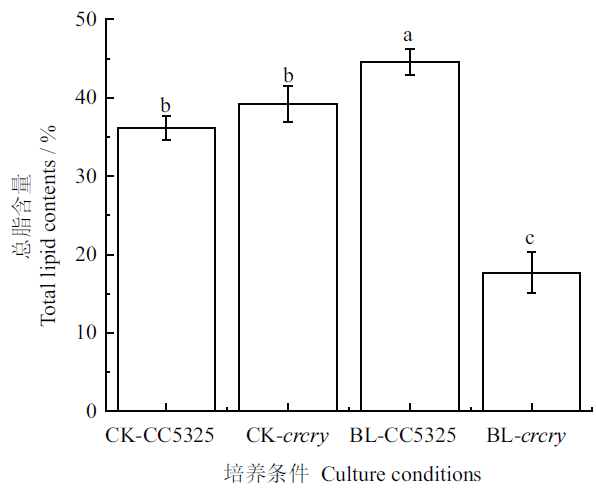
Fig. 5 Effects of blue light on the total lipid of C. Reinh-ardtii CC5325 and crcry Different lowercase letters indicate significant difference at 0.05 level
| [1] |
崔红利, 陈军, 侯义龙, 等. 真核微藻蓝光受体及其功能研究进展[J]. 生物技术通报, 2017, 33(4): 51-62.
doi: 10.13560/j.cnki.biotech.bull.1985.2017.04.007 |
| Cui HL, Chen J, Hou YL, et al. Research progress on blue-photoreceptors and its functions in eukaryotic microalgae[J]. Biotechnol Bull, 2017, 33(4): 51-62. | |
| [2] |
Butler WL, Norris KH, Siegelman HW, et al. Detection, assay, and preliminary purification of the pigment controlling photoresponsive development of plants[J]. Proc Natl Acad Sci USA, 1959, 45(12): 1703-1708.
pmid: 16590561 |
| [3] | Chen J, Kong Y, Wang Z, et al. Negative phototropism of Chlorophytum comosum roots and their mechanisms[J]. Hortic Plant J, 2015, 1(1): 55-60. |
| [4] |
Chen J, Mo YW, Xu HW. Calcium signaling is involved in negative phototropism of rice seminal roots[J]. Rice Sci, 2014, 21(1): 39-46.
doi: 10.1016/S1672-6308(13)60162-6 |
| [5] |
Galvão VC, Fankhauser C. Sensing the light environment in plants: photoreceptors and early signaling steps[J]. Curr Opin Neurobiol, 2015, 34: 46-53.
doi: 10.1016/j.conb.2015.01.013 pmid: 25638281 |
| [6] |
Sakai T, Kagawa T, Kasahara M, et al. Arabidopsis nph1 and npl1: blue light receptors that mediate both phototropism and chloroplast relocation[J]. Proc Natl Acad Sci USA, 2001, 98(12): 6969-6974.
doi: 10.1073/pnas.101137598 pmid: 11371609 |
| [7] | Garner WW, Allard HA. Effect of the relative length of day and night and other factors of the environment on growth and reproduction in plants[J]. Mon Wea Rev, 1920, 48(7): 415. |
| [8] | 闫海芳, 周波, 李玉花. 光受体及光信号转导[J]. 植物学通报, 2004, 39(2): 235-246. |
| Yan HF, Zhou B, Li YH. Photoreceptors and light signal transduction[J]. Chin Bull Bot, 2004, 39(2): 235-246. | |
| [9] |
Lepetit B, Dietzel L. Light signaling in photosynthetic eukaryotes with ‘green’ and ‘red’ chloroplasts[J]. Environ Exp Bot, 2015, 114: 30-47.
doi: 10.1016/j.envexpbot.2014.07.007 URL |
| [10] |
Cashmore AR. The cryptochrome family of photoreceptors[J]. Plant Cell Environ, 1997, 20(6): 764-767.
doi: 10.1046/j.1365-3040.1997.d01-125.x URL |
| [11] |
Lin CT, Shalitin D. Cryptochrome structure and signal transduction[J]. Annu Rev Plant Biol, 2003, 54: 469-496.
pmid: 14503000 |
| [12] |
Sancar A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors[J]. Chem Rev, 2003, 103(6): 2203-2237.
pmid: 12797829 |
| [13] |
Lin CT, Shalitin D. Cryptochrome structure and signal transduction[J]. Annu Rev Plant Biol, 2003, 54: 469-496.
pmid: 14503000 |
| [14] |
Shalitin D, Yu XH, Maymon M, et al. Blue light-dependent in vivo and in vitro phosphorylation of Arabidopsis cryptochrome 1[J]. Plant Cell, 2003, 15(10): 2421-2429.
doi: 10.1105/tpc.013011 pmid: 14523249 |
| [15] |
Kianianmomeni A, Hallmann A. Algal photoreceptors: in vivo functions and potential applications[J]. Planta, 2014, 239(1): 1-26.
doi: 10.1007/s00425-013-1962-5 pmid: 24081482 |
| [16] | 孙燕, 许志茹. 植物的蓝光受体[J]. 植物生理学通讯, 2008, 44(1): 144-150. |
| Sun Y, Xu ZR. Blue-light photoreceptors in plant[J]. Plant Physiol Commun, 2008, 44(1): 144-150. | |
| [17] |
Chaves I, Pokorny R, Byrdin M, et al. The cryptochromes: blue light photoreceptors in plants and animals[J]. Annu Rev Plant Biol, 2011, 62: 335-364.
doi: 10.1146/annurev-arplant-042110-103759 pmid: 21526969 |
| [18] |
Coesel S, Mangogna M, Ishikawa T, et al. Diatom PtCPF1 is a new cryptochrome/photolyase family member with DNA repair and transcription regulation activity[J]. EMBO Rep, 2009, 10(6): 655-661.
doi: 10.1038/embor.2009.59 pmid: 19424294 |
| [19] |
Cock JM, Sterck L, Rouzé P, et al. The Ectocarpus genome and the independent evolution of multicellularity in brown algae[J]. Nature, 2010, 465(7298): 617-621.
doi: 10.1038/nature09016 URL |
| [20] |
Beel B, Müller N, Kottke T, et al. News about cryptochrome photoreceptors in algae[J]. Plant Signal Behav, 2013, 8(2): e22870.
doi: 10.4161/psb.22870 URL |
| [21] |
Heijde M, Zabulon G, Corellou F, et al. Characterization of two members of the cryptochrome/photolyase family from Ostreococcus tauri provides insights into the origin and evolution of cryptochromes[J]. Plant Cell Environ, 2010, 33(10): 1614-1626.
doi: 10.1111/j.1365-3040.2010.02168.x URL |
| [22] |
Asimgil H, Kavakli IH. Purification and characterization of five members of photolyase/cryptochrome family from Cyanidioschyzon merolae[J]. Plant Sci, 2012, 185/186: 190-198.
doi: 10.1016/j.plantsci.2011.10.005 URL |
| [23] |
Oliveri P, Fortunato AE, Petrone L, et al. The cryptochrome/photolyase family in aquatic organisms[J]. Mar Genomics, 2014, 14: 23-37.
doi: 10.1016/j.margen.2014.02.001 URL |
| [24] |
Immeln D, Schlesinger R, Heberle J, et al. Blue light induces radical formation and autophosphorylation in the light-sensitive domain of Chlamydomonas cryptochrome[J]. J Biol Chem, 2007, 282(30): 21720-21728.
doi: 10.1074/jbc.M700849200 URL |
| [25] |
Immeln D, Pokorny R, Herman E, et al. Photoreaction of plant and DASH cryptochromes probed by infrared spectroscopy: the neutral radical state of flavoproteins[J]. J Phys Chem B, 2010, 114(51): 17155-17161.
doi: 10.1021/jp1076388 URL |
| [26] |
Beel B, Prager K, Spexard M, et al. A flavin binding cryptochrome photoreceptor responds to both blue and red light in Chlamydomonas reinhardtii[J]. Plant Cell, 2012, 24(7): 2992-3008.
doi: 10.1105/tpc.112.098947 URL |
| [27] |
Langenbacher T, Immeln D, Dick B, et al. Microsecond light-induced proton transfer to flavin in the blue light sensor plant cryptochrome[J]. J Am Chem Soc, 2009, 131(40): 14274-14280.
doi: 10.1021/ja901628y pmid: 19754110 |
| [28] |
Li XB, Zhang R, Patena W, et al. An indexed, mapped mutant library enables reverse genetics studies of biological processes in Chlamydomonas reinhardtii[J]. Plant Cell, 2016, 28(2): 367-387.
doi: 10.1105/tpc.15.00465 URL |
| [29] |
Zhang R, Patena W, Armbruster U, et al. High-throughput genotyping of green algal mutants reveals random distribution of mutagenic insertion sites and endonucleolytic cleavage of transforming DNA[J]. Plant Cell, 2014, 26(4): 1398-1409.
doi: 10.1105/tpc.114.124099 URL |
| [30] |
Ahmed RA, He ML, Aftab RA, et al. Bioenergy application of Dunaliella salina SA 134 grown at various salinity levels for lipid production[J]. Sci Rep, 2017, 7(1): 8118.
doi: 10.1038/s41598-017-07540-x URL |
| [31] | 邵雪梅. 不同氮浓度对埃氏小球藻生长及油脂积累的影响[D]. 太谷: 山西农业大学, 2016. |
| Shao XM. Effects of different nitrogen concentrations on growth and lipid accumulation of Chlorella emersionii[D]. Taigu: Shanxi Agricultural University, 2016. | |
| [32] |
Ninu L, Ahmad M, Miarelli C, et al. Cryptochrome 1 controls tomato development in response to blue light[J]. Plant J, 1999, 18(5): 551-556.
doi: 10.1046/j.1365-313X.1999.00466.x URL |
| [33] |
Imaizumi T, Kadota A, Hasebe M, et al. Cryptochrome light signals control development to suppress auxin sensitivity in the moss Physcomitrella patens[J]. Plant Cell, 2002, 14(2): 373-386.
pmid: 11884681 |
| [34] |
Kanegae T, Wada M. Isolation and characterization of homologues of plant blue-light photoreceptor(cryptochrome)genes from the fern Adiantum capillus-veneris[J]. Mol Gen Genet, 1998, 259(4): 345-353.
doi: 10.1007/s004380050821 URL |
| [35] |
Das P, Lei W, Aziz SS, et al. Enhanced algae growth in both phototrophic and mixotrophic culture under blue light[J]. Bioresour Technol, 2011, 102(4): 3883-3887.
doi: 10.1016/j.biortech.2010.11.102 URL |
| [36] |
Gonçalves VD, Fagundes-Klen MR, Trigueros DEG, et al. Combination of light emitting diodes(LEDs)for photostimulation of carotenoids and chlorophylls synthesis in Tetradesmus sp[J]. Algal Res, 2019, 43: 101649.
doi: 10.1016/j.algal.2019.101649 URL |
| [37] |
Kuwano K, Abe N, Nishi Y, et al. Growth and cell cycle of Ulva compressa(Ulvophyceae)under LED illumination[J]. J Phycol, 2014, 50(4): 744-752.
doi: 10.1111/jpy.12207 URL |
| [38] | 沈银武, 朱运芝, 刘永定. 不同光质对中华植生藻的影响[J]. 水生生物学报, 1999, 23(3): 285-287. |
| Shen YW, Zhu YZ, Liu YD. Effects of different light quality on richelia sinica[J]. Acta Hydrobiol Sin, 1999, 23(3): 285-287. | |
| [39] |
Im CS, Eberhard S, Huang KY, et al. Phototropin involvement in the expression of genes encoding chlorophyll and carotenoid biosynthesis enzymes and LHC apoproteins in Chlamydomonas reinhardtii[J]. Plant J, 2006, 48(1): 1-16.
doi: 10.1111/j.1365-313X.2006.02852.x URL |
| [40] | 高松, 刘颖, 刘学娜, 等. 光质对大葱叶片碳氮代谢的影响[J]. 植物生理学报, 2020, 56(3): 565-572. |
| Gao S, Liu Y, Liu XN, et al. Effects of light quality on carbon and nitrogen metabolism in leaves of Welsh onion(Allium fistulosum)[J]. Plant Physiol J, 2020, 56(3): 565-572. | |
| [41] | 李尚, 陶益, 刀国华, 等. 紫外线对再生水中斜生栅藻的生长抑制效果[J]. 环境工程, 2020, 38(10): 97-102, 113. |
| Li S, Tao Y, Dao GH, et al. Growth suppression effect of UV-C irradiation on Scenedesmus obliquus in reclaimed water[J]. Environ Eng, 2020, 38(10): 97-102, 113. | |
| [42] | 周玉娇, 李亚军, 费小雯, 等. 小球藻紫外线诱变及高含油藻株筛选[J]. 热带作物学报, 2010, 31(12): 2124-2129. |
| Zhou YJ, Li YJ, Fei XW, et al. UV-irradiation of Chlorella vulgaris and screening of petroliferous strains[J]. Chin J Trop Crops, 2010, 31(12): 2124-2129. | |
| [43] |
Atta M, Idris A, Bukhari A, et al. Intensity of blue LED light: a potential stimulus for biomass and lipid content in fresh water microalgae Chlorella vulgaris[J]. Bioresour Technol, 2013, 148: 373-378.
doi: 10.1016/j.biortech.2013.08.162 URL |
| [44] |
Fortunato AE, Jaubert M, Enomoto G, et al. Diatom phytochromes reveal the existence of far-red-light-based sensing in the ocean[J]. Plant Cell, 2016, 28(3): 616-628.
doi: 10.1105/tpc.15.00928 URL |
| [45] |
Ma RJ, Thomas-Hall SR, Chua ET, et al. Gene expression profiling of astaxanthin and fatty acid pathways in Haematococcus pluvialis in response to different LED lighting conditions[J]. Bioresour Technol, 2018, 250: 591-602.
doi: 10.1016/j.biortech.2017.11.094 URL |
| [1] | CHEN Guang, LI Jia, DU Rui-ying, WANG Xu. Identification and Gene Functional Analysis of Salinity-hypersensitive Mutant ss2 in Rice [J]. Biotechnology Bulletin, 2022, 38(9): 158-166. |
| [2] | TANG Guang-fu, GUI Yan-ling, MAN Hai-qiao, ZHAO Jie-hong. Editing pyrG Gene of Monascus by CRISPR/Cas 9 and Its Effects on Secondary Metabolism [J]. Biotechnology Bulletin, 2022, 38(8): 198-205. |
| [3] | ZHOU Shi-chen, YI Zhi-ben, WANG Xin-yi, YANG Xiao-ying, SUN Li-na, LUAN Wei-jiang, LIANG Shan-shan. Genetic Analysis and Gene Mapping of Sorghum Double-grain Mutant Dgs [J]. Biotechnology Bulletin, 2022, 38(7): 171-177. |
| [4] | TANG Yue-hui, ZHAO Yu-fan, LIN Jin, WANG Yin, CAO Bo-yuan, CHE Yi-fan, YANG Wen-jie, BAO Xin-xin, YANG Tong-wen. Identification and Gene Mapping of a Seedling Lethal Mutant in Rice [J]. Biotechnology Bulletin, 2022, 38(10): 124-131. |
| [5] | FAN Yu-chen, LU Yao, LIU Xiang-nan, ZHAO Bo. Construction of Mutants Swapping Ubiquitin E3 Ligase CHIP and E4B U-box Domain and Verification of Ubiquitination Activity [J]. Biotechnology Bulletin, 2021, 37(12): 191-197. |
| [6] | ZHU Cai-lin, LÜ Xiang, XIA Xiao-le. Effect of Site-directed Mutagenesis of Amino Acids in Lid Region on the Enzymatic Properties of T1 Lipase [J]. Biotechnology Bulletin, 2020, 36(11): 94-102. |
| [7] | BI Yan-zhen, XIAO Hong-wei, ZHANG Li-ping, REN Hong-yan, HUA Zai-dong, HUA Wen-jun, WANG Zheng, NIU Min-jie, LIN Zheng-yun, REN Xi-dong, SUN Li-hua, ZHENG Xin-min. Applications and Challenges of Gene Editing in Non-human Primate Models [J]. Biotechnology Bulletin, 2018, 34(5): 48-56. |
| [8] | LI Su-zhen, YANG Wen-zhu, CHEN Ru-mei. An Overview on Yellow Green Leaf Mutants in Rice [J]. Biotechnology Bulletin, 2018, 34(11): 15-21. |
| [9] | WANG Wen-xiu, WANG Lei. Research Progress on Maize Dwarf Genes [J]. Biotechnology Bulletin, 2018, 34(11): 22-26. |
| [10] | LIU Qi-ming,CHEN Zhen-hua,HAN Xiao. Monosaccharide Components of the Male Sterile Mutant gsl5 in Rice [J]. Biotechnology Bulletin, 2017, 33(9): 116-119. |
| [11] | WANG Jia-mei, AN Xue-jiao, ZHANG Zhi-guo. Mapping-based Cloning of a Wax Crystal-Sparse Leaf Mutant wcl1 in Rice [J]. Biotechnology Bulletin, 2017, 33(8): 46-50. |
| [12] | LIU Xiao-li, JIANG Shi-jie, XUE Dong, LIU Ying-ying, WU Xiao-li, FENG Shuai, HAN Jia-hui, WANG Yu-zhou, PING Shu-zhen, WANG Jin. Construction and Biological Characterization of Gene dlp Deletion Mutant of Deinococcus radiodurans R1 [J]. Biotechnology Bulletin, 2017, 33(2): 155-163. |
| [13] | CHEN Yun1, NIU Chun-qing1, SONG Xiao-shuang1, SU Chang1, HUA Zi-chun2, LIU Yan1. Expression and Activity Analysis of DSPAα1 Deletion Mutants in Pichia pastoris [J]. Biotechnology Bulletin, 2016, 32(9): 246-252. |
| [14] | LI Jian-yu,LI Bo,CHEN Hua-min,YANG Feng-huan,HE Chen-yang,TIAN Fang. Identification of Critical Residues in c-di-GMP Receptor Clpxoo from Xanthomonas oryzae pv. oryzae [J]. Biotechnology Bulletin, 2016, 32(12): 124-129. |
| [15] | Zhang Longdi, Wang Yanwei, Zhang Zhiguo, Wu Jinxia. Genetic Analysis and Gene Mapping of a Dominant Rolled Leaf Mutant z2 in Rice [J]. Biotechnology Bulletin, 2015, 31(6): 100-105. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||