








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (5): 63-76.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1006
Previous Articles Next Articles
MA Fang-fang( ), LIU Guan-wen, PANG Bing, JIANG Chun-mei, SHI Jun-ling(
), LIU Guan-wen, PANG Bing, JIANG Chun-mei, SHI Jun-ling( )
)
Received:2022-08-19
Online:2023-05-26
Published:2023-06-08
MA Fang-fang, LIU Guan-wen, PANG Bing, JIANG Chun-mei, SHI Jun-ling. Strategies of Increasing Flavonoid Production in Engineered Bacteria by Intensifying the Efflux of Flavonoid in Cells[J]. Biotechnology Bulletin, 2023, 39(5): 63-76.
| 基因 Gene | 转运蛋白Transporter | 位置/转运方向 Location/Transport direction | 类黄酮 Flavonoid | 基因来源 Source of gene | 工程菌 Engineering bacterium | 参考文献 Reference |
|---|---|---|---|---|---|---|
| Abcc1 (NC_000082) | MRP | 小肠基底外侧膜/到血液 | 柚皮素Naringin 橙皮素Hesperidin (葡萄糖醛酸化和甲基化Glucuronidation and methylation) | 小家鼠 Mus musculus(House mouse) | 大肠杆菌Escherichia coli 金黄色葡萄糖球菌Staphylococcus aureus | [ |
| Abcc2 (NC_000085) | MRP | 小肠细胞顶点/到小肠 | 柚皮素Naringin 橙皮素Hesperidin (葡萄糖醛酸化和甲基化Glucuronidation and methylation) | 小家鼠 Mus musculus(House mouse) | 苏云金芽孢杆菌Bacillus thuringiensis | [ |
| Abcc3 (NC_000077) | MRP | 小肠基底外侧膜/到血液 | 柚皮素Naringin (葡萄糖醛酸化和甲基化Glucuronidation and methylation) | 小家鼠 Mus musculus(House mouse) | 大肠杆菌Escherichia coli 金黄色葡萄糖球菌Staphylococcus aureus | [ |
| MRP2 (NC_000017) | MRP | 小肠基底外侧膜/到血液 | 表没食子儿茶素没食子酸酯Epigallocatechin gallate(硫酸化和甲基化Sulfation and methylation) | 人类 Homo sapiens(Human) | 酿酒酵母Saccharomyces cerevisiae | [ |
| ABCB1 (NC_000007) | P-gp | 肠道管腔细胞/参与外排 | 4'-羟-6-甲基黄酮 4'-hydroxy-6- Methylflavone 4'-羟-6-甲氧基黄酮 4'-hydroxy-6-methoxyflavone 4'-羟-β-萘基黄酮 4'-hydroxy-β- Naphthoflavone | 人类 Homo sapiens(Human) | 金黄色葡萄糖球菌Staphylococcus aureus MRSA6 | [ |
| ABCG2 (NC_000072) | BCRP | 肠道管腔细胞/参与外排 | 染料木素Genistein (硫酸化和葡萄糖醛酸化Sulfation and glucuronidation) | 小家鼠 Mus musculus(House mouse) | 大肠杆菌Escherichia coli | [ |
Table 1 Animal flavonoid transport genes successfully expressed in genetically engineered bacteria
| 基因 Gene | 转运蛋白Transporter | 位置/转运方向 Location/Transport direction | 类黄酮 Flavonoid | 基因来源 Source of gene | 工程菌 Engineering bacterium | 参考文献 Reference |
|---|---|---|---|---|---|---|
| Abcc1 (NC_000082) | MRP | 小肠基底外侧膜/到血液 | 柚皮素Naringin 橙皮素Hesperidin (葡萄糖醛酸化和甲基化Glucuronidation and methylation) | 小家鼠 Mus musculus(House mouse) | 大肠杆菌Escherichia coli 金黄色葡萄糖球菌Staphylococcus aureus | [ |
| Abcc2 (NC_000085) | MRP | 小肠细胞顶点/到小肠 | 柚皮素Naringin 橙皮素Hesperidin (葡萄糖醛酸化和甲基化Glucuronidation and methylation) | 小家鼠 Mus musculus(House mouse) | 苏云金芽孢杆菌Bacillus thuringiensis | [ |
| Abcc3 (NC_000077) | MRP | 小肠基底外侧膜/到血液 | 柚皮素Naringin (葡萄糖醛酸化和甲基化Glucuronidation and methylation) | 小家鼠 Mus musculus(House mouse) | 大肠杆菌Escherichia coli 金黄色葡萄糖球菌Staphylococcus aureus | [ |
| MRP2 (NC_000017) | MRP | 小肠基底外侧膜/到血液 | 表没食子儿茶素没食子酸酯Epigallocatechin gallate(硫酸化和甲基化Sulfation and methylation) | 人类 Homo sapiens(Human) | 酿酒酵母Saccharomyces cerevisiae | [ |
| ABCB1 (NC_000007) | P-gp | 肠道管腔细胞/参与外排 | 4'-羟-6-甲基黄酮 4'-hydroxy-6- Methylflavone 4'-羟-6-甲氧基黄酮 4'-hydroxy-6-methoxyflavone 4'-羟-β-萘基黄酮 4'-hydroxy-β- Naphthoflavone | 人类 Homo sapiens(Human) | 金黄色葡萄糖球菌Staphylococcus aureus MRSA6 | [ |
| ABCG2 (NC_000072) | BCRP | 肠道管腔细胞/参与外排 | 染料木素Genistein (硫酸化和葡萄糖醛酸化Sulfation and glucuronidation) | 小家鼠 Mus musculus(House mouse) | 大肠杆菌Escherichia coli | [ |
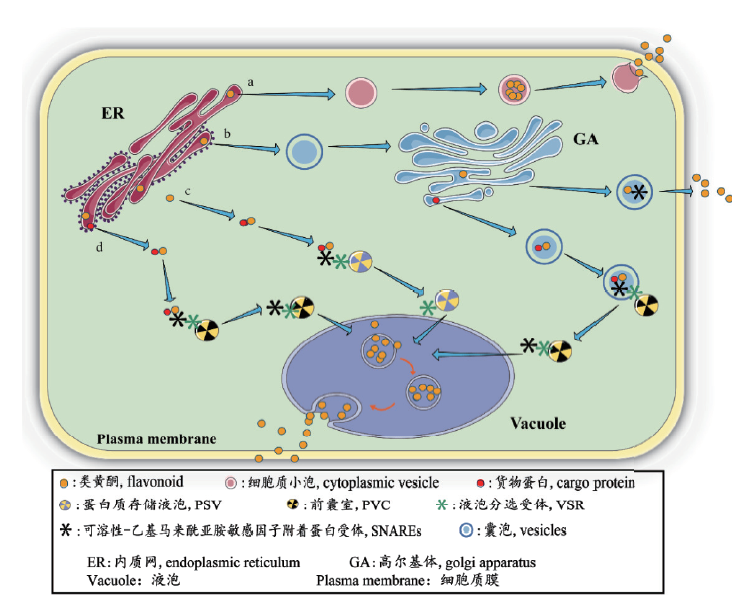
Fig. 1 Vesicle-mediated flavonoid transport in plants a:Tapetal corpuscles transport flavonoids by means of cytoplasmic vesicles; b:vesicle fusion transport is mediated by vacuolar sorting receptors(VSR)and SNARE in general plants; c:arabidopsis uses the protein storage vacuole to enter the central vacuole; d:anthocyanins enter the central large vacuole by means of cargo protein fusion anterior chamber
| 基因 Gene | 转运蛋白 Transporter | 转运底物 Substrate | 分布位置 Location | 基因来源 Gene source | 工程菌Engineering microorganism | 参考文献 Reference |
|---|---|---|---|---|---|---|
| TRANSPARENT TESTA12(tt12) (AJ294464) | MATE | 花青素-3-葡萄糖Cyanidin-3-O-glucoside | 花青素苷液泡Anthocyanin vacuole | 拟南芥Arabidopsis thaliana | 大肠杆菌Escherichia coli酿酒酵母Saccharomyces cerevisiae | [ |
| 花青素-3-葡萄糖转移酶 Cyanidin 3-O-Glucosyltransferase(3GT) (NC_003076) | UDP-d-葡萄糖转运蛋白 UDP-d-glucose transporter proteins GLF | 花青素-3-葡萄糖Cyanidin-3-O-glucoside | 细胞质Cytoplasm | 拟南芥Arabidopsis thaliana | 大肠杆菌Escherichia coli BL21 | [ |
| 花青素-3-葡萄糖转移酶 Cyanidin 3-O-Glucosyltransferase(3GT) (NC_003076) | UDP-d-葡萄糖转运蛋白 UDP-d-glucose transporter proteins GALU1 | 花青素-3-葡萄糖Cyanidin-3-O-glucoside | 细胞质Cytoplasm | 拟南芥Arabidopsis thaliana | 谷氨酸棒状杆菌Corynebacterium glutamicum | [ |
| ZmMrp3 (AY609318) | MRP | 花青素-3-葡萄糖Cyanidin-3-O-glucoside | 花青素苷液泡Anthocyanin vacuole | 玉米 Zea mays | 大肠杆菌Escherichia coli | [ |
| Bronze-2(Bz2) (NC_050096) | GST | 花青素-3-葡萄糖Cyanidin-3-O-glucoside | 细胞质Cytoplasm | 玉米 Zea mays | 大肠杆菌Escherichia coli | [ |
| 葡萄原花青素O-甲基转移酶Vitis vinifera Anthocyanin O-methyltransferase(VvAOMT) (NC_012007) | 花色素苷O-甲基转移酶Anthocyanin O-methyltransferase(AOMT) | 花青素-3-葡萄糖Cyanidin 3-O-glucoside, 甲基花青素3-O-葡萄糖 Peonidin 3-O- glucoside | 细胞质Cytoplasm | 葡萄 Vitis vinifera | 大肠杆菌Escherichia coli DH5α and BL21 | [ |
Table 2 Plant flavonoid transporters successfully expressed in genetically engineered bacteria
| 基因 Gene | 转运蛋白 Transporter | 转运底物 Substrate | 分布位置 Location | 基因来源 Gene source | 工程菌Engineering microorganism | 参考文献 Reference |
|---|---|---|---|---|---|---|
| TRANSPARENT TESTA12(tt12) (AJ294464) | MATE | 花青素-3-葡萄糖Cyanidin-3-O-glucoside | 花青素苷液泡Anthocyanin vacuole | 拟南芥Arabidopsis thaliana | 大肠杆菌Escherichia coli酿酒酵母Saccharomyces cerevisiae | [ |
| 花青素-3-葡萄糖转移酶 Cyanidin 3-O-Glucosyltransferase(3GT) (NC_003076) | UDP-d-葡萄糖转运蛋白 UDP-d-glucose transporter proteins GLF | 花青素-3-葡萄糖Cyanidin-3-O-glucoside | 细胞质Cytoplasm | 拟南芥Arabidopsis thaliana | 大肠杆菌Escherichia coli BL21 | [ |
| 花青素-3-葡萄糖转移酶 Cyanidin 3-O-Glucosyltransferase(3GT) (NC_003076) | UDP-d-葡萄糖转运蛋白 UDP-d-glucose transporter proteins GALU1 | 花青素-3-葡萄糖Cyanidin-3-O-glucoside | 细胞质Cytoplasm | 拟南芥Arabidopsis thaliana | 谷氨酸棒状杆菌Corynebacterium glutamicum | [ |
| ZmMrp3 (AY609318) | MRP | 花青素-3-葡萄糖Cyanidin-3-O-glucoside | 花青素苷液泡Anthocyanin vacuole | 玉米 Zea mays | 大肠杆菌Escherichia coli | [ |
| Bronze-2(Bz2) (NC_050096) | GST | 花青素-3-葡萄糖Cyanidin-3-O-glucoside | 细胞质Cytoplasm | 玉米 Zea mays | 大肠杆菌Escherichia coli | [ |
| 葡萄原花青素O-甲基转移酶Vitis vinifera Anthocyanin O-methyltransferase(VvAOMT) (NC_012007) | 花色素苷O-甲基转移酶Anthocyanin O-methyltransferase(AOMT) | 花青素-3-葡萄糖Cyanidin 3-O-glucoside, 甲基花青素3-O-葡萄糖 Peonidin 3-O- glucoside | 细胞质Cytoplasm | 葡萄 Vitis vinifera | 大肠杆菌Escherichia coli DH5α and BL21 | [ |
| 类黄酮 Flavonoid | 外排基因 Efflux gene | 微生物 Microorganism | 转运蛋白家族 Transporter protein | 参考文献 Reference |
|---|---|---|---|---|
| 查尔酮Chalcone (2E)-1-(4'-氨苯基)-3-(4-甲氧苯基)- 2丙-烯-1-酮 (2E)-1-(4'-aminophenyl)-3-(4-methoxyphenyl)-prop-2-en-1-one | MepA (NC_016941) | 金黄色葡萄球菌Staphylococcus aureus | ABC转运蛋白 ABC transporters | [ |
| 木犀草素Luteolin 芹菜素Apigenin 山奈酚Kaempferol 鼠李素Rhamnetin | NorA (NC_007795) | 金黄色葡萄球菌Staphylococcus aureus NCTC 8325 | ABC转运蛋白 ABC transporters | [ |
| 染料木素Genistein 芹菜素Apigenin 表儿茶素没食子酸酯Epicatechin gallate | NorA、TetK、MsrA(NC_007795) | 金黄色葡萄球菌Staphylococcus aureus NCTC 8325 | ABC转运蛋白 ABC transporters | [ |
| 二氢黄酮Flavonone | LmrA (NZ_CP059048) | 乳酸乳球菌 Lactococcus lactis | ABC转运蛋白 ABC transporters | [ |
| 类黄酮Flavonoid | YvcC (AGE65090) | 枯草芽孢杆菌 Bacillus subtilis | ABC转运蛋白 ABC transporters | [ |
| 类黄酮Flavonoid | DrrAB (AAC43341) | 波赛链霉菌 Streptomyces peucetius | ABC转运蛋白 ABC transporters | [ |
| 原儿茶酸Protocatechuic-acid 龙胆酸Gentisic-acid 毛花甙丙Lanatoside-C 大豆甙元Diadzein | MexB (NC_002516) | 铜绿假单胞菌Pseudomonas aeruginosa PAO1 | RND转运蛋白 RND transporters | [ |
| 龙胆酸Gentisic-acid 毛花甙丙Lanatoside-C 大豆甙元Diadzein | AcrB (NC_000913) | 大肠杆菌 Escherichia coli str. K-12 substr. MG1655 | RND转运蛋白 RND transporters | [ |
| 表没食子儿茶素没食子酸酯Epigallocatechin gallate | CmeABC、CmeDEF (NC_002163) | 空肠弯曲菌Campylobacter jejuni NCTC 11168 | RND转运蛋白 RND transporters | [ |
| 槲皮素Quercetin | AcrAB-TolC (NC_000913) | 大肠杆菌 Escherichia coli str. K-12 substr. MG1655 | RND转运蛋白 RND transporters | [ |
| 硫酸原黄素Proflavine | Acel (QWB10945) | 鲍曼不动杆菌Acinetobacter baumannii | PACE转运蛋白 PACE transporters | [ |
Table 3 Microbial transporters involved in flavonoid efflux
| 类黄酮 Flavonoid | 外排基因 Efflux gene | 微生物 Microorganism | 转运蛋白家族 Transporter protein | 参考文献 Reference |
|---|---|---|---|---|
| 查尔酮Chalcone (2E)-1-(4'-氨苯基)-3-(4-甲氧苯基)- 2丙-烯-1-酮 (2E)-1-(4'-aminophenyl)-3-(4-methoxyphenyl)-prop-2-en-1-one | MepA (NC_016941) | 金黄色葡萄球菌Staphylococcus aureus | ABC转运蛋白 ABC transporters | [ |
| 木犀草素Luteolin 芹菜素Apigenin 山奈酚Kaempferol 鼠李素Rhamnetin | NorA (NC_007795) | 金黄色葡萄球菌Staphylococcus aureus NCTC 8325 | ABC转运蛋白 ABC transporters | [ |
| 染料木素Genistein 芹菜素Apigenin 表儿茶素没食子酸酯Epicatechin gallate | NorA、TetK、MsrA(NC_007795) | 金黄色葡萄球菌Staphylococcus aureus NCTC 8325 | ABC转运蛋白 ABC transporters | [ |
| 二氢黄酮Flavonone | LmrA (NZ_CP059048) | 乳酸乳球菌 Lactococcus lactis | ABC转运蛋白 ABC transporters | [ |
| 类黄酮Flavonoid | YvcC (AGE65090) | 枯草芽孢杆菌 Bacillus subtilis | ABC转运蛋白 ABC transporters | [ |
| 类黄酮Flavonoid | DrrAB (AAC43341) | 波赛链霉菌 Streptomyces peucetius | ABC转运蛋白 ABC transporters | [ |
| 原儿茶酸Protocatechuic-acid 龙胆酸Gentisic-acid 毛花甙丙Lanatoside-C 大豆甙元Diadzein | MexB (NC_002516) | 铜绿假单胞菌Pseudomonas aeruginosa PAO1 | RND转运蛋白 RND transporters | [ |
| 龙胆酸Gentisic-acid 毛花甙丙Lanatoside-C 大豆甙元Diadzein | AcrB (NC_000913) | 大肠杆菌 Escherichia coli str. K-12 substr. MG1655 | RND转运蛋白 RND transporters | [ |
| 表没食子儿茶素没食子酸酯Epigallocatechin gallate | CmeABC、CmeDEF (NC_002163) | 空肠弯曲菌Campylobacter jejuni NCTC 11168 | RND转运蛋白 RND transporters | [ |
| 槲皮素Quercetin | AcrAB-TolC (NC_000913) | 大肠杆菌 Escherichia coli str. K-12 substr. MG1655 | RND转运蛋白 RND transporters | [ |
| 硫酸原黄素Proflavine | Acel (QWB10945) | 鲍曼不动杆菌Acinetobacter baumannii | PACE转运蛋白 PACE transporters | [ |
| 微生物 Microorganism | 修饰 Modification | 酶类 Enzyme | 修饰位点 Modification site | 参考文献Reference |
|---|---|---|---|---|
| 乳酸菌Lactobacillus 乳球菌Lactococcus 双歧杆菌Bifidobacteria 肠球菌Enterococcus 链霉菌Streptomyces 黑曲霉Aspergillus niger | 糖基化Glycosylation | 糖基转移酶 Glycosyltransferase 糖苷酶Glycosidase 淀粉蔗糖酶Starch sucrase α-淀粉酶α -amylase | 主要为B环C-3'羟基,然后依次为B环C-4'羟基> A环C-7> A环C-5> A环C-8 | [ |
| 南极假丝酵母 Candida antarctica | 乙酰基化Acetoxylation | 脂肪酶Lipase 蛋白酶Protease | 主要为伯羟基,酰基化程度取决于酰基供体脂肪酸、芳香酸和乙烯基酯的链长 | [ |
| 灰色链霉菌Streptomyces griseus 球孢白僵菌Beauveria bassiana 枯草芽孢杆菌Bacillus subtilis | 羟基化Hydroxylation | 酪氨酸酶Tyrosinase | 主要为B环C-3'羟基 | [ |
| 球孢白僵菌Beauveria bassiana 大肠杆菌Escherichia coli 链霉菌KCTC 0041BP Streptomyces sp. KCTC 0041BP 灰色链霉菌Streptomyces griseus | 甲基化Methylation | 甲基转移酶 Methyltransferase | 主要为B环C-3'和C-4'羟基,然后依次为A环C-7 >A环C-5 >C环 C-3 | [ |
Table 4 Enzymes that catalyze flavonoid derivatives in microorganisms
| 微生物 Microorganism | 修饰 Modification | 酶类 Enzyme | 修饰位点 Modification site | 参考文献Reference |
|---|---|---|---|---|
| 乳酸菌Lactobacillus 乳球菌Lactococcus 双歧杆菌Bifidobacteria 肠球菌Enterococcus 链霉菌Streptomyces 黑曲霉Aspergillus niger | 糖基化Glycosylation | 糖基转移酶 Glycosyltransferase 糖苷酶Glycosidase 淀粉蔗糖酶Starch sucrase α-淀粉酶α -amylase | 主要为B环C-3'羟基,然后依次为B环C-4'羟基> A环C-7> A环C-5> A环C-8 | [ |
| 南极假丝酵母 Candida antarctica | 乙酰基化Acetoxylation | 脂肪酶Lipase 蛋白酶Protease | 主要为伯羟基,酰基化程度取决于酰基供体脂肪酸、芳香酸和乙烯基酯的链长 | [ |
| 灰色链霉菌Streptomyces griseus 球孢白僵菌Beauveria bassiana 枯草芽孢杆菌Bacillus subtilis | 羟基化Hydroxylation | 酪氨酸酶Tyrosinase | 主要为B环C-3'羟基 | [ |
| 球孢白僵菌Beauveria bassiana 大肠杆菌Escherichia coli 链霉菌KCTC 0041BP Streptomyces sp. KCTC 0041BP 灰色链霉菌Streptomyces griseus | 甲基化Methylation | 甲基转移酶 Methyltransferase | 主要为B环C-3'和C-4'羟基,然后依次为A环C-7 >A环C-5 >C环 C-3 | [ |
| [1] |
Rauf A, Imran M, Khan IA, et al. Anticancer potential of quercetin: a comprehensive review[J]. Phytother Res, 2018, 32(11): 2109-2130.
doi: 10.1002/ptr.6155 pmid: 30039547 |
| [2] |
Cao QM, Yan JY, Sun ZC, et al. Simultaneous optimization of ultrasound-assisted extraction for total flavonoid content and antioxidant activity of the tender stem of Triarrhena lutarioriparia using response surface methodology[J]. Food Sci Biotechnol, 2021, 30(1): 37-45.
doi: 10.1007/s10068-020-00851-2 |
| [3] |
Liang JP, Li WQ, Jia XY, et al. Transcriptome sequencing and characterization of Astragalus membranaceus var. mongholicus root reveals key genes involved in flavonoids biosynthesis[J]. Genes Genomics, 2020, 42(8): 901-914.
doi: 10.1007/s13258-020-00953-5 |
| [4] |
Lu Y, Che JX, Xu XG, et al. Metabolomics reveals the response of the phenylpropanoid biosynthesis pathway to starvation treatment in the grape endophyte Alternaria sp. MG1[J]. J Agric Food Chem, 2020, 68(4): 1126-1135.
doi: 10.1021/acs.jafc.9b05302 URL |
| [5] |
Lu Y, Ye C, Che JX, et al. Genomic sequencing, genome-scale metabolic network reconstruction, and in silico flux analysis of the grape endophytic fungus Alternaria sp. MG1[J]. Microb Cell Fact, 2019, 18(1): 13.
doi: 10.1186/s12934-019-1063-7 |
| [6] |
Álvarez-Álvarez R, Botas A, Albillos SM, et al. Molecular genetics of naringenin biosynthesis, a typical plant secondary metabolite produced by Streptomyces clavuligerus[J]. Microb Cell Fact, 2015, 14: 178.
doi: 10.1186/s12934-015-0373-7 pmid: 26553209 |
| [7] |
Delmulle T, de Maeseneire SL, de Mey M. Challenges in the microbial production of flavonoids[J]. Phytochem Rev, 2018, 17(2): 229-247.
doi: 10.1007/s11101-017-9515-3 URL |
| [8] |
Lu Y, Shao DY, Shi JL, et al. Strategies for enhancing resveratrol production and the expression of pathway enzymes[J]. Appl Microbiol Biotechnol, 2016, 100(17): 7407-7421.
doi: 10.1007/s00253-016-7723-1 pmid: 27405437 |
| [9] |
Che J, Shi J, Gao Z, et al. A new approach to produce resveratrol by enzymatic bioconversion[J]. J Microbiol Biotechnol, 2016, 26(8): 1348-1357.
doi: 10.4014/jmb.1512.12084 URL |
| [10] | Dymarska M, Janeczko T, Kostrzewa-Susłow E. Glycosylation of methoxylated flavonoids in the cultures of Isaria fumosorosea KCH J2[J]. Molecules, 2018, 23(10): E2578. |
| [11] |
Dymarska M, Janeczko T, Kostrzewa-Susłow E. Glycosylation of 3-hydroxyflavone, 3-methoxyflavone, quercetin and baicalein in fungal cultures of the genus Isaria[J]. Molecules, 2018, 23(10): 2477.
doi: 10.3390/molecules23102477 URL |
| [12] |
Gutmann H, Hruz P, Zimmermann C, et al. Distribution of breast cancer resistance protein(BCRP/ABCG2)mRNA expression along the human GI tract[J]. Biochem Pharmacol, 2005, 70(5): 695-699.
pmid: 15998509 |
| [13] |
Rich GT, Buchweitz M, Winterbone MS, et al. Towards an understanding of the low bioavailability of quercetin: a study of its interaction with intestinal lipids[J]. Nutrients, 2017, 9(2): 111.
doi: 10.3390/nu9020111 URL |
| [14] |
Fang YJ, Liang FQ, Liu KY, et al. Structure characteristics for intestinal uptake of flavonoids in Caco-2 cells[J]. Food Res Int, 2018, 105: 353-360.
doi: S0963-9969(17)30818-9 pmid: 29433224 |
| [15] |
Zhang L, Zheng Y, Chow MSS, et al. Investigation of intestinal absorption and disposition of green tea catechins by Caco-2 monolayer model[J]. Int J Pharm, 2004, 287(1-2): 1-12.
doi: 10.1016/j.ijpharm.2004.08.020 URL |
| [16] |
Lespine A, Dupuy J, Orlowski S, et al. Interaction of ivermectin with multidrug resistance proteins(MRP1, 2 and 3)[J]. Chem Biol Interact, 2006, 159(3): 169-179.
doi: 10.1016/j.cbi.2005.11.002 URL |
| [17] |
Jiang YK, Jiang ZY, Ma L, et al. Advances in nanodelivery of green tea catechins to enhance the anticancer activity[J]. Molecules, 2021, 26(11): 3301.
doi: 10.3390/molecules26113301 URL |
| [18] |
El-Ashmawy NE, El-Zamarany EA, Khedr EG, et al. Effect of modification of MTDH gene expression on colorectal cancer aggressiveness[J]. Gene, 2019, 698: 92-99.
doi: S0378-1119(19)30203-3 pmid: 30836117 |
| [19] |
Zhang L, Lin G, Kovács B, et al. Mechanistic study on the intestinal absorption and disposition of baicalein[J]. Eur J Pharm Sci, 2007, 31(3-4): 221-231.
pmid: 17507208 |
| [20] |
Duan JZ, Xie Y, Luo HL, et al. Transport characteristics of isorhamnetin across intestinal Caco-2 cell monolayers and the effects of transporters on it[J]. Food Chem Toxicol, 2014, 66: 313-320.
doi: 10.1016/j.fct.2014.02.003 pmid: 24525098 |
| [21] |
Chung JO, Lee SB, Jeong KH, et al. Quercetin and fisetin enhanced the small intestine cellular uptake and plasma levels of epi-catechins in in vitro and in vivo models[J]. Food Funct, 2018, 9(1): 234-242.
doi: 10.1039/C7FO01576C URL |
| [22] |
Chen ZL, Ma TT, Huang C, et al. Efficiency of transcellular transport and efflux of flavonoids with different glycosidic units from flavonoids of Litsea coreana L. in a MDCK epithelial cell monolayer model[J]. Eur J Pharm Sci, 2014, 53: 69-76.
doi: 10.1016/j.ejps.2013.12.010 URL |
| [23] | 吴桐, 刘晓东. 参与P-gp膜定位和功能调控的相关蛋白研究进展[J]. 药学进展, 2021, 45(2): 130-136. |
| Wu T, Liu XD. Research progress of proteins involved in the location and function regulation of P-gp on plasma membrane[J]. Prog Pharm Sci, 2021, 45(2): 130-136. | |
| [24] |
Kanado Y, Tsurudome Y, Omata Y, et al. Estradiol regulation of P-glycoprotein expression in mouse kidney and human tubular epithelial cells, implication for renal clearance of drugs[J]. Biochem Biophys Res Commun, 2019, 519(3): 613-619.
doi: 10.1016/j.bbrc.2019.09.021 URL |
| [25] |
Mohos V, Fliszár-Nyúl E, Ungvári O, et al. Inhibitory effects of quercetin and its main methyl, sulfate, and glucuronic acid conjugates on cytochrome P450 enzymes, and on OATP, BCRP and MRP2 transporters[J]. Nutrients, 2020, 12(8): 2306.
doi: 10.3390/nu12082306 URL |
| [26] |
Liu YY, Lu YY, Li XY, et al. Kaempferol suppression of acute colitis is regulated by the efflux transporters BCRP and MRP2[J]. Eur J Pharm Sci, 2022, 179: 106303.
doi: 10.1016/j.ejps.2022.106303 URL |
| [27] |
Pei SH, Dou YY, Zhang WK, et al. O-Sulfation disposition of curcumin and quercetin in SULT1A3 overexpressing HEK293 cells: the role of arylsulfatase B in cellular O-sulfation regulated by transporters[J]. Food Funct, 2022, 13(20): 10558-10573.
doi: 10.1039/D2FO01436J URL |
| [28] |
Qu FF, Ai ZY, Liu SY, et al. Study on mechanism of low bioavailability of black tea theaflavins by using Caco-2 cell monolayer[J]. Drug Deliv, 2021, 28(1): 1737-1747.
doi: 10.1080/10717544.2021.1949074 pmid: 34463173 |
| [29] |
Guo X, Cao XD, Fang XG, et al. Involvement of phase II enzymes and efflux transporters in the metabolism and absorption of naringin, hesperidin and their aglycones in rats[J]. Int J Food Sci Nutr, 2022, 73(4): 480-490.
doi: 10.1080/09637486.2021.2012562 URL |
| [30] |
Swartz TH, Ito M, Ohira T, et al. Catalytic properties of Staphylococcus aureus and Bacillus members of the secondary cation/proton antiporter-3(Mrp)family are revealed by an optimized assay in an Escherichia coli host[J]. J Bacteriol, 2007, 189(8): 3081-3090.
pmid: 17293423 |
| [31] |
Ma YM, Zhang JF, Xiao YT, et al. The cadherin Cry1Ac binding-region is necessary for the cooperative effect with ABCC2 transporter enhancing insecticidal activity of Bacillus thuringiensis Cry1Ac toxin[J]. Toxins, 2019, 11(9): 538.
doi: 10.3390/toxins11090538 URL |
| [32] |
Kikuchi T, Hayashi A, Ikeda N, et al. Multidrug resistance-associated protein 2(MRP2)is an efflux transporter of EGCG and its metabolites in the human small intestine[J]. J Nutr Biochem, 2022, 107: 109071.
doi: 10.1016/j.jnutbio.2022.109071 URL |
| [33] |
Nouemsi GRS, Jouda JB, Leutcha PB, et al. A new flavonol derivative and other compounds from the leaves of Bauhinia thonningii Schum with activity against multidrug-resistant bacteria[J]. Nat Prod Res, 2022, 29(2): 1-9.
doi: 10.1080/14786419.2014.955488 URL |
| [34] |
Dankó B, Tóth S, Martins A, et al. Synthesis and SAR study of anticancer protoflavone derivatives: investigation of cytotoxicity and interaction with ABCB1 and ABCG2 multidrug efflux transporters[J]. ChemMedChem, 2017, 12(11): 850-859.
doi: 10.1002/cmdc.201700225 pmid: 28436164 |
| [35] |
Koirala N, Pandey RP, Thuan NH, et al. Metabolic engineering ofEscherichia colifor the production of isoflavonoid-4’-O-methoxides and their biological activities[J]. Biotechnol Appl Biochem, 2019, 66(4): 484-493.
doi: 10.1002/bab.v66.4 URL |
| [36] |
Zhu W, Xu HY, Wang SWJ, et al. Breast cancer resistance protein(BCRP)and sulfotransferases contribute significantly to the disposition of genistein in mouse intestine[J]. AAPS J, 2010, 12(4): 525-536.
doi: 10.1208/s12248-010-9209-x URL |
| [37] |
Gomez C, Conejero G, Torregrosa L, et al. In vivo grapevine anthocyanin transport involves vesicle-mediated trafficking and the contribution of anthoMATE transporters and GST[J]. Plant J, 2011, 67(6): 960-970.
doi: 10.1111/tpj.2011.67.issue-6 URL |
| [38] |
Zhao Y, Dong WQ, Zhu YC, et al. PpGST1, an anthocyanin-related glutathione S-transferase gene, is essential for fruit coloration in peach[J]. Plant Biotechnol J, 2020, 18(5): 1284-1295.
doi: 10.1111/pbi.13291 pmid: 31693790 |
| [39] |
DaSilva LLP, Foresti O, Denecke J. Targeting of the plant vacuolar sorting receptor BP80 is dependent on multiple sorting signals in the cytosolic tail[J]. Plant Cell, 2006, 18(6): 1477-1497.
doi: 10.1105/tpc.105.040394 pmid: 16714388 |
| [40] |
Poustka F, Irani NG, Feller A, et al. A trafficking pathway for anthocyanins overlaps with the endoplasmic Reticulum-to-vacuole protein-sorting route in Arabidopsis and contributes to the formation of vacuolar inclusions[J]. Plant Physiol, 2007, 145(4): 1323-1335.
doi: 10.1104/pp.107.105064 URL |
| [41] | Dean JV, Willis M, Shaban L. Transport of acylated anthocyanins by the Arabidopsis ATP-binding cassette transporters AtABCC1, AtABCC2, and AtABCC14[J]. Physiol Plant, 2022, 174(5): e13780. |
| [42] |
Hsieh K, Huang AHC. Tapetosomes in Brassica tapetum accumulate endoplasmic Reticulum-derived flavonoids and alkanes for delivery to the pollen surface[J]. Plant Cell, 2007, 19(2): 582-596.
pmid: 17307923 |
| [43] |
Zhang HB, Wang L, Deroles S, et al. New insight into the structures and formation of anthocyanic vacuolar inclusions in flower petals[J]. BMC Plant Biol, 2006, 6: 29.
pmid: 17173704 |
| [44] |
Takanashi K, Shitan N, Yazaki K. The multidrug and toxic compound extrusion(MATE)family in plants[J]. Plant Biotechnol, 2014, 31(5): 417-430.
doi: 10.5511/plantbiotechnology.14.0904a URL |
| [45] |
He X, Szewczyk P, Karyakin A, et al. Structure of a cation-bound multidrug and toxic compound extrusion transporter[J]. Nature, 2010, 467(7318): 991-994.
doi: 10.1038/nature09408 |
| [46] |
Upadhyay N, Kar D, Deepak Mahajan B, et al. The multitasking abilities of MATE transporters in plants[J]. J Exp Bot, 2019, 70(18): 4643-4656.
doi: 10.1093/jxb/erz246 pmid: 31106838 |
| [47] |
Kitamura S, Oono Y, Narumi I. Arabidopsis pab1, a mutant with reduced anthocyanins in immature seeds from banyuls, harbors a mutation in the MATE transporter FFT[J]. Plant Mol Biol, 2016, 90(1-2): 7-18.
doi: 10.1007/s11103-015-0389-8 pmid: 26608698 |
| [48] |
Xu H, Yang PP, Cao YW, et al. Cloning and functional characterization of a flavonoid transport-related MATE gene in Asiatic hybrid lilies(Lilium spp.)[J]. Genes, 2020, 11(4): 418.
doi: 10.3390/genes11040418 URL |
| [49] |
李跃, 李国瑞, 陈永胜. 微生物代谢工程在花色苷生产过程中的应用现状和前景[J]. 食品科学, 2020, 41(13): 260-266.
doi: 10.7506/spkx1002-6630-20190618-202 |
| Li Y, Li GR, Chen YS. Present and future application of microbial metabolic engineering in anthocyanin production[J]. Food Sci, 2020, 41(13): 260-266. | |
| [50] |
Kallscheuer N, Vogt M, Bott M, et al. Functional expression of plant-derived O-methyltransferase, flavanone 3-hydroxylase, and flavonol synthase in Corynebacterium glutamicum for production of pterostilbene, kaempferol, and quercetin[J]. J Biotechnol, 2017, 258: 190-196.
doi: S0168-1656(17)30023-8 pmid: 28143765 |
| [51] |
Marinova K, Pourcel L, Weder B, et al. The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H+-antiporter active in proanthocyanidin-accumulating cells of the seed coat[J]. Plant Cell, 2007, 19(6): 2023-2038.
doi: 10.1105/tpc.106.046029 URL |
| [52] |
Debeaujon I, Peeters AJ, Léon-Kloosterziel KM, et al. The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium[J]. Plant Cell, 2001, 13(4): 853-871.
doi: 10.1105/tpc.13.4.853 pmid: 11283341 |
| [53] |
Shrestha B, Pandey RP, Darsandhari S, et al. Combinatorial approach for improved cyanidin 3-O-glucoside production in Escherichia coli[J]. Microb Cell Fact, 2019, 18(1): 7.
doi: 10.1186/s12934-019-1056-6 pmid: 30654816 |
| [54] |
Zha J, Zang Y, Mattozzi M, et al. Metabolic engineering of Corynebacterium glutamicum for anthocyanin production[J]. Microb Cell Fact, 2018, 17: 143.
doi: 10.1186/s12934-018-0990-z |
| [55] |
Goodman CD, Casati P, Walbot V. A multidrug resistance-associated protein involved in anthocyanin transport in Zea mays[J]. Plant Cell, 2004, 16(7): 1812-1826.
doi: 10.1105/tpc.022574 URL |
| [56] |
Marrs KA, Alfenito MR, Lloyd AM, et al. A glutathione S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2[J]. Nature, 1995, 375(6530): 397-400.
doi: 10.1038/375397a0 |
| [57] |
Cress BF, Leitz QD, Kim DC, et al. CRISPRi-mediated metabolic engineering of E. coli for O-methylated anthocyanin production[J]. Microb Cell Fact, 2017, 16(1): 10.
doi: 10.1186/s12934-016-0623-3 |
| [58] |
Waditzer M, Bucar F. Flavonoids as inhibitors of bacterial efflux pumps[J]. Molecules, 2021, 26(22): 6904.
doi: 10.3390/molecules26226904 URL |
| [59] | Le MT, Trinh DTT, Ngo TD, et al. Chalcone derivatives as potential inhibitors of P-glycoprotein and NorA: an in silico and in vitro study[J]. Biomed Res Int, 2022, 2022: 9982453. |
| [60] | Freitas TS, Xavier JC, Pereira RLS, et al. In vitro and in silico studies of chalcones derived from natural acetophenone inhibitors of NorA and MepA multidrug efflux pumps in Staphylococcus aureus[J]. Microb Pathog, 2021, 161(Pt B): 105286. |
| [61] |
Alves Borges Leal AL, Teixeira da Silva P, Nunes da Rocha M, et al. Potentiating activity of Norfloxacin by synthetic chalcones against NorA overproducing Staphylococcus aureus[J]. Microb Pathog, 2021, 155: 104894.
doi: 10.1016/j.micpath.2021.104894 URL |
| [62] |
Brown AR, Ettefagh KA, Todd D, et al. A mass spectrometry-based assay for improved quantitative measurements of efflux pump inhibition[J]. PLoS One, 2015, 10(5): e0124814.
doi: 10.1371/journal.pone.0124814 URL |
| [63] |
陈福暖, 黄瑜, 蔡佳, 等. ABC转运蛋白结构及其在细菌致病性中的研究进展[J]. 生物技术通报, 2022, 38(6): 43-52.
doi: 10.13560/j.cnki.biotech.bull.1985.2021-1175 |
| Chen FN, Huang Y, Cai J, et al. Structure of ABC transporter and research progress of it in bacterial pathogenicity[J]. Biotechnol Bull, 2022, 38(6): 43-52. | |
| [64] |
Wang SY, Sun ZL, Liu T, et al. Flavonoids from Sophora moorcroftiana and their synergistic antibacterial effects on MRSA[J]. Phytother Res, 2014, 28(7): 1071-1076.
doi: 10.1002/ptr.5098 URL |
| [65] |
Guo YR, Huang CC, Su HY, et al. Luteolin increases susceptibility to macrolides by inhibiting MsrA efflux pump in Trueperella pyogenes[J]. Vet Res, 2022, 53(1): 3.
doi: 10.1186/s13567-021-01021-w |
| [66] |
van Veen HW, Venema K, Bolhuis H, et al. Multidrug resistance mediated by a bacterial homolog of the human multidrug transporter MDR1[J]. Proc Natl Acad Sci USA, 1996, 93(20): 10668-10672.
pmid: 8855237 |
| [67] |
Steinfels E, Orelle C, Dalmas O, et al. Highly efficient over-production in E. coli of YvcC, a multidrug-like ATP-binding cassette transporter from Bacillus subtilis[J]. Biochim Biophys Acta Biomembr, 2002, 1565(1): 1-5.
doi: 10.1016/S0005-2736(02)00515-1 URL |
| [68] |
Guilfoile PG, Hutchinson CR. A bacterial analog of the mdr gene of mammalian tumor cells is present in Streptomyces peucetius, the producer of daunorubicin and doxorubicin[J]. PNAS, 1991, 88(19): 8553-8557.
pmid: 1924314 |
| [69] |
Li XZ, Plésiat P, Nikaido H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria[J]. Clin Microbiol Rev, 2015, 28(2): 337-418.
doi: 10.1128/CMR.00117-14 URL |
| [70] |
Aparna V, Dineshkumar K, Mohanalakshmi N, et al. Identification of natural compound inhibitors for multidrug efflux pumps of Escherichia coli and Pseudomonas aeruginosa using in silico high-throughput virtual screening and in vitro validation[J]. PLoS One, 2014, 9(7): e101840.
doi: 10.1371/journal.pone.0101840 URL |
| [71] |
Plé C, Tam HK, Vieira da Cruz A, et al. Pyridylpiperazine-based allosteric inhibitors of RND-type multidrug efflux pumps[J]. Nat Commun, 2022, 13(1): 115.
doi: 10.1038/s41467-021-27726-2 pmid: 35013254 |
| [72] |
Kurinčič M, Klančnik A, Smole Možina S. Epigallocatechin gallate as a modulator of Campylobacter resistance to macrolide antibiotics[J]. Int J Antimicrob Agents, 2012, 40(5): 467-471.
doi: 10.1016/j.ijantimicag.2012.07.015 URL |
| [73] |
Chen MY, Shi XD, Yu ZL, et al. In situ structure of the AcrAB-TolC efflux pump at subnanometer resolution[J]. Structure, 2022, 30(1): 107-113.
doi: 10.1016/j.str.2021.08.008 URL |
| [74] |
Ohene-Agyei T, Mowla R, Rahman T, et al. Phytochemicals increase the antibacterial activity of antibiotics by acting on a drug efflux pump[J]. MicrobiologyOpen, 2014, 3(6): 885-896.
doi: 10.1002/mbo3.212 pmid: 25224951 |
| [75] | Hassan KA, Liu Q, Henderson PJF, et al. Homologs of the Acinetobacter baumannii AceI transporter represent a new family of bacterial multidrug efflux systems[J]. mBio, 2015, 6(1): e01982-e01914. |
| [76] |
Khemthong S, Nuonming P, Dokpikul T, et al. Regulation and function of the flavonoid-inducible efflux system, emrR-emrAB, in Agrobacterium tumefaciens C58[J]. Appl Microbiol Biotechnol, 2019, 103(14): 5763-5780.
doi: 10.1007/s00253-019-09899-5 |
| [77] |
Garrido-Arandia M, Garrido-Arandia M, Silva-Navas J, et al. Characterisation of a flavonoid ligand of the fungal protein alta1[J]. Sci Rep, 2016, 6: 33468.
doi: 10.1038/srep33468 pmid: 27633190 |
| [78] |
Cao H, Chen XQ, Jassbi AR, et al. Microbial biotransformation of bioactive flavonoids[J]. Biotechnol Adv, 2015, 33(1): 214-223.
doi: S0734-9750(14)00161-X pmid: 25447420 |
| [79] |
Slana M, Zigon D, Makovec T, et al. The response of filamentous fungus Rhizopus nigricans to flavonoids[J]. J Basic Microbiol, 2011, 51(4): 433-441.
doi: 10.1002/jobm.v51.4 URL |
| [80] |
Hosny M, Dhar K, Rosazza JP. Hydroxylations and methylations of quercetin, fisetin, and catechin by Streptomyces griseus[J]. J Nat Prod, 2001, 64(4): 462-465.
doi: 10.1021/np000457m pmid: 11325228 |
| [81] | Che JX, Shi JL, Gao ZH, et al. Transcriptome analysis reveals the genetic basis of the resveratrol biosynthesis pathway in an endophytic fungus(Alternaria sp. MG1)isolated from Vitis vinifera[J]. Front Microbiol, 2016, 7: 1257. |
| [82] |
Zhu J, Yan L, Xu XG, et al. Strategies to enhance the production of pinoresinol and its glucosides by endophytic fungus(Phomopsis sp. XP-8)isolated from Tu-chung bark[J]. AMB Express, 2018, 8(1): 55.
doi: 10.1186/s13568-018-0584-5 |
| [83] |
Wang Q, Teng MK, Li X. Functional and structural characterization of a novel catechol-O-methyltransferase from Schizosaccharomyces pombe[J]. IUBMB Life, 2019, 71(3): 330-339.
doi: 10.1002/iub.v71.3 URL |
| [84] |
Liu XN, Cheng J, Zhu XX, et al. De novo biosynthesis of multiple pinocembrin derivatives in Saccharomyces cerevisiae[J]. ACS Synth Biol, 2020, 9(11): 3042-3051.
doi: 10.1021/acssynbio.0c00289 URL |
| [85] |
Zhou JW, Wang K, Xu S, et al. Identification of membrane proteins associated with phenylpropanoid tolerance and transport in Escherichia coli BL21[J]. J Proteomics, 2015, 113: 15-28.
doi: 10.1016/j.jprot.2014.09.012 URL |
| [86] |
Li JH, Tian CF, Xia YH, et al. Production of plant-specific flavones baicalein and scutellarein in an engineered E. coli from available phenylalanine and tyrosine[J]. Metab Eng, 2019, 52: 124-133.
doi: 10.1016/j.ymben.2018.11.008 URL |
| [87] |
Solopova A, van Tilburg AY, Foito A, et al. Engineering Lactococcus lactis for the production of unusual anthocyanins using tea as substrate[J]. Metab Eng, 2019, 54: 160-169.
doi: S1096-7176(19)30059-X pmid: 30978503 |
| [88] |
Abari AH, Tayebi M. Bioconversion of genistein to orobol by Bacillus subtilis spore displayed tyrosinase and monitoring the anticancer effects of orobol on MCF-7 breast cancer cells[J]. Biotechnol Bioprocess Eng, 2019, 24(3): 507-512.
doi: 10.1007/s12257-019-0067-9 |
| [89] |
Gao S, Lyu YB, Zeng WZ, et al. Efficient biosynthesis of(2S)-naringenin from p-coumaric acid in Saccharomyces cerevisiae[J]. J Agric Food Chem, 2020, 68(4): 1015-1021.
doi: 10.1021/acs.jafc.9b05218 URL |
| [90] |
Gao S, Zhou HR, Zhou JW, et al. Promoter-library-based pathway optimization for efficient(2S)-naringenin production from p-coumaric acid in Saccharomyces cerevisiae[J]. J Agric Food Chem, 2020, 68(25): 6884-6891.
doi: 10.1021/acs.jafc.0c01130 URL |
| [91] |
Lyu X, Zhao G, Ng KR, et al. Metabolic engineering of Saccharo-myces cerevisiae for de novo production of kaempferol[J]. J Agric Food Chem, 2019, 67(19): 5596-5606.
doi: 10.1021/acs.jafc.9b01329 URL |
| [92] |
Li G, Li H, Lyu Y, et al. Enhanced biosynthesis of dihydromyricetin in Saccharomyces cerevisiae by coexpression of multiple hydroxylases[J]. J Agric Food Chem, 2020, 68(48): 14221-14229.
doi: 10.1021/acs.jafc.0c05261 URL |
| [93] |
Akram M, Rasool A, An T, et al. Metabolic engineering of Yarrowia lipolytica for liquiritigenin production[J]. Chem Eng Sci, 2021, 230: 116177.
doi: 10.1016/j.ces.2020.116177 URL |
| [94] |
Marín L, Gutiérrez-Del-Río I, Entrialgo-Cadierno R, et al. De novo biosynthesis of myricetin, kaempferol and quercetin in Streptomyces albus and Streptomyces coelicolor[J]. PLoS One, 2018, 13(11): e0207278.
doi: 10.1371/journal.pone.0207278 URL |
| [95] |
Marín L, Gutiérrez-Del-Río I, Yagüe P, et al. De novo biosynthesis of apigenin, luteolin, and eriodictyol in the actinomycete Streptomyces albus and production improvement by feeding and spore conditioning[J]. Front Microbiol, 2017, 8: 921.
doi: 10.3389/fmicb.2017.00921 URL |
| [96] |
Sun JC, Sun WT, Zhang GL, et al. High efficient production of plant flavonoids by microbial cell factories: challenges and opportunities[J]. Metab Eng, 2022, 70: 143-154.
doi: 10.1016/j.ymben.2022.01.011 URL |
| [97] |
Bai HW, Shim JY, Yu JN, et al. Biochemical and molecular modeling studies of the O-methylation of various endogenous and exogenous catechol substrates catalyzed by recombinant human soluble and membrane-bound catechol-O-methyltransferases[J]. Chem Res Toxicol, 2007, 20(10): 1409-1425.
doi: 10.1021/tx700174w URL |
| [98] |
Byeon Y, Lee HY, Lee K, et al. Caffeic acid O-methyltransferase is involved in the synthesis of melatonin by methylating N-acetylserotonin in Arabidopsis[J]. J Pineal Res, 2014, 57(2): 219-227.
doi: 10.1111/jpi.12160 URL |
| [99] |
Byeon Y, Choi GH, Lee HY, et al. Melatonin biosynthesis requires N-acetylserotonin methyltransferase activity of caffeic acid O-methyltransferase in rice[J]. J Exp Bot, 2015, 66(21): 6917-6925.
doi: 10.1093/jxb/erv396 URL |
| [100] |
Zhang YF, He YZ, Zhang N, et al. Combining protein and metabolic engineering strategies for biosynthesis of melatonin in Escherichia coli[J]. Microb Cell Fact, 2021, 20(1): 170.
doi: 10.1186/s12934-021-01662-8 |
| [101] |
Chen ZY, Shen XL, Wang J, et al. Establishing an artificial pathway for de novo biosynthesis of vanillyl alcohol in Escherichia coli[J]. ACS Synth Biol, 2017, 6(9): 1784-1792.
doi: 10.1021/acssynbio.7b00129 URL |
| [102] |
Rodrigues JL, Gomes D, Rodrigues LR. A combinatorial approach to optimize the production of curcuminoids from tyrosine in Escherichia coli[J]. Front Bioeng Biotechnol, 2020, 8: 59.
doi: 10.3389/fbioe.2020.00059 URL |
| [103] |
Heo KT, Kang SY, Hong YS. De novo biosynthesis of pterostilbene in an Escherichia coli strain using a new resveratrol O-methyltransferase from Arabidopsis[J]. Microb Cell Fact, 2017, 16(1): 30.
doi: 10.1186/s12934-017-0644-6 URL |
| [104] |
Guelfo JR, Rodríguez-Rojas A, Matic I, et al. A MATE-family efflux pump rescues the Escherichia coli 8-oxoguanine-repair-deficient mutator phenotype and protects against H2O2 killing[J]. PLoS Genet, 2010, 6(5): e1000931.
doi: 10.1371/journal.pgen.1000931 URL |
| [105] |
Long F, Rouquette-Loughlin C, Shafer WM, et al. Functional cloning and characterization of the multidrug efflux pumps NorM from Neisseria gonorrhoeae and YdhE from Escherichia coli[J]. Antimicrob Agents Chemother, 2008, 52(9): 3052-3060.
doi: 10.1128/AAC.00475-08 URL |
| [106] |
Magar RT, Sohng JK. A review on structure, modifications and structure-activity relation of quercetin and its derivatives[J]. J Microbiol Biotechnol, 2020, 30(1): 11-20.
doi: 10.4014/jmb.1907.07003 URL |
| [107] |
Gaya P, Peirotén Á, Medina M, et al. Isoflavone metabolism by a collection of lactic acid bacteria and bifidobacteria with biotechnological interest[J]. Int J Food Sci Nutr, 2016, 67(2): 117-124.
doi: 10.3109/09637486.2016.1144724 pmid: 26878882 |
| [108] |
Ma BJ, Zeng J, Shao L, et al. Efficient bioconversion of quercetin into a novel glycoside by Streptomyces rimosus subsp. rimosus ATCC 10970[J]. J Biosci Bioeng, 2013, 115(1): 24-26.
doi: 10.1016/j.jbiosc.2012.07.020 URL |
| [109] |
Ye HY, Li XJ, Li LY, et al. Homologous expression and characterization of α-L-rhamnosidase from Aspergillus niger for the transformation of flavonoids[J]. Appl Biochem Biotechnol, 2022, 194(8): 3453-3467.
doi: 10.1007/s12010-022-03894-9 |
| [110] |
Hao X, Xie JH, Li YT, et al. Acetylated pelargonidin-3- O-glucoside exhibits promising thermostability, lipophilicity, and protectivity against oxidative damage by activating the Nrf2/ARE pathway[J]. Food Funct, 2022, 13(5): 2618-2630.
doi: 10.1039/D2FO00179A URL |
| [111] |
Fernandez-Aulis F, Torres A, Sanchez-Mendoza E, et al. New acylated cyanidin glycosides extracted from underutilized potential sources: Enzymatic synthesis, antioxidant activity and thermostability[J]. Food Chem, 2020, 309: 125796.
doi: 10.1016/j.foodchem.2019.125796 URL |
| [112] | de M B Costa EM, Pimenta FC, Luz WC, et al. Selection of filamentous fungi of the Beauveria genus able to metabolize quercetin like mammalian cells[J]. Braz J Microbiol, 2008, 39(2): 405-408. |
| [113] |
Darsandhari S, Dhakal D, Shrestha B, et al. Characterization of regioselective flavonoid O-methyltransferase from the Streptomyces sp. KCTC 0041BP[J]. Enzyme Microb Technol, 2018, 113: 29-36.
doi: 10.1016/j.enzmictec.2018.02.007 URL |
| [114] |
Chang TS, Chao SY, Chen YC. Production of ortho-hydroxydaidzein derivatives by a recombinant strain of Pichia pastoris harboring a cytochrome P450 fusion gene[J]. Process Biochem, 2013, 48(3): 426-429.
doi: 10.1016/j.procbio.2013.02.014 URL |
| [1] | ZHANG He-chen, YUAN Xin, GAO Jie, WANG Xiao-chen, WANG Hui-juan, LI Yan-min, WANG Li-min, FU Zhen-zhu, LI Bao-yin. Mechanism of Flower Petal Coloration and Molecular Breeding [J]. Biotechnology Bulletin, 2023, 39(5): 23-31. |
| [2] | DUAN Yue-tong, WANG Peng-nian, ZHANG Chun-bao, LIN Chun-jing. Research Progress in Plant Flavanone-3-hydroxylase Gene [J]. Biotechnology Bulletin, 2022, 38(6): 27-33. |
| [3] | YAO Yu, GU Jia-jun, SUN Chao, SHEN Guo-an, GUO Bao-lin. Advances in Plant Flavonoids UDP-glycosyltransferase [J]. Biotechnology Bulletin, 2022, 38(12): 47-57. |
| [4] | LUO Ya-fang, ZHU Chun-hua, XIAO Yu-ting, LI Fang-quan, ZHANG Jiang, WANG Yu-shu. Screening and Functional Analysis of UGT Genes Involved in the Flavonoid Biosynthesis of Brassica oleracea var. acephala [J]. Biotechnology Bulletin, 2022, 38(11): 194-201. |
| [5] | YE Min, GAO Jiao-qi, ZHOU Yong-jin. Engineering Non-conventional Yeast Cell Factory for the Biosynthesis of Natural Products [J]. Biotechnology Bulletin, 2021, 37(8): 12-24. |
| [6] | WANG Jun-yi, DONG Jin-jin, LIU Wei, CAO Fu-liang, WANG Gui-bin, WANG Yi-qiang. Research on the Growth,Browning and Flavonoid Accumulation of Ginkgo biloba Callus [J]. Biotechnology Bulletin, 2019, 35(2): 16-22. |
| [7] | YANG Fei-yun, BAI Jie, LIU Kun, WANG Rui-gang. Increase of Flavonoids Content in Arabidopsis Heterologous Expressing CiCHI Gene [J]. Biotechnology Bulletin, 2019, 35(11): 39-45. |
| [8] | LI Ping ,ZHANG Gui-ping, HU Jian-ran. Effects of Total Flavonoids from Forsythia suspense on the Proliferation of Gastric Cancer Cell MGC80-3 [J]. Biotechnology Bulletin, 2018, 34(6): 199-203. |
| [9] | TAO Bo FANG Mei ZHANG Jia-nan OU Yun-wen JIA Ning. Determination and Purification of Total Flavonoids from Seeds of Ammopiptanthus mongolica [J]. Biotechnology Bulletin, 2017, 33(5): 63-70. |
| [10] | SHEN Xiao-lin, YUAN Qi-peng. Research Advance on Biosynthesis of Aromatic Amino Acids and Their Derivatives [J]. Biotechnology Bulletin, 2017, 33(1): 24-34. |
| [11] | ZHAO Jing, LI Nan, WU Ru, YANG Zhan-wei, HU Wen-bing, WANG Wen-jun. Research Advance on the Effect of Food Functional Components on Animal Genomic DNA Methylation [J]. Biotechnology Bulletin, 2016, 32(1): 15-19. |
| [12] | Liu Xiaodan, Zhang Keqin, Liu Lian, Li Wenchang. Determination the Content of Total Flavonoids and Hypericin in Callus from Hypericum attenuatum Choisy [J]. Biotechnology Bulletin, 2015, 31(1): 98-103. |
| [13] | Liu Shan, Sun Rong, Tang Zizhong, Gao Jinglei, Hu Hongli, Chen Hui. Genetic Diversity of Wild Populations of Conyza blinii Lévl. and Extraction of Total Flavonoids [J]. Biotechnology Bulletin, 2013, 0(9): 84-88. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||