








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (7): 195-205.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1304
Previous Articles Next Articles
YU Hui( ), WANG Jing, LIANG Xin-xin, XIN Ya-ping, ZHOU Jun, ZHAO Hui-jun(
), WANG Jing, LIANG Xin-xin, XIN Ya-ping, ZHOU Jun, ZHAO Hui-jun( )
)
Received:2022-10-24
Online:2023-07-26
Published:2023-08-17
Contact:
ZHAO Hui-jun
E-mail:1632570918@qq.com;zhaohuijun1022@163.com
YU Hui, WANG Jing, LIANG Xin-xin, XIN Ya-ping, ZHOU Jun, ZHAO Hui-jun. Isolation and Functional Verification of Genes Responding to Iron and Cadmium Stresses in Lycium barbarum[J]. Biotechnology Bulletin, 2023, 39(7): 195-205.
| 引物Primer | 序列Sequence(5'-3') | 用途Purpose |
|---|---|---|
| LbIRT2 | F: ATGGCTGCTCAAACCCAAGT R: TCACAACTCCATGCCAGTAGTAGA | 基因克隆 Gene clone |
| LbFRO | F: ATGGGTGAACTCTCAGGGCAA R: CTAGAGGTCAAAATTGTGGCAGTTG | 基因克隆 Gene clone |
| LbNRAMP3 | F: ATGCCTCCACACGATGAAGAACA R: TCAACTCTCTATGCTCGTGATACTCTT | 基因克隆 Gene clone |
| LbActin | F: TCTACGAGGGTTACGCTTTG R: TCCCGTTCAGCAGTGGTT | 枸杞内参引物 Internal primer of Lycium barbarum L. |
| qRT-LbFRO | F: TCAAGCCACCCAGGGAACT R: CATGAAAATTGGTAAACTGAACATCA | 荧光定量引物 Fluorescent quantitative PCR primer |
| qRT-LbNRAMP3 | F: GCAGAGAGCGTTAATAACAAGAAGTTG R: TTAAGCCACTCGTTGAGAACATCT | 荧光定量引物 Fluorescent quantitative PCR primer |
| qRT-LbIRT1 | F: AGGAGCAATTCATCAAGATGCA R: TGCTACATGCTCCGGCTACTAG | 荧光定量引物 Fluorescent quantitative PCR primer |
| qRT-LbIRT2 | F: TCCAAATTACCATTCCTAAAGACCAT R: CGAACCCACTGTGATCTTTGAG | 荧光定量引物 Fluorescent quantitative PCR primer |
| qRT-LbYSL | F: TGCAGTGTTCATGAGCGGTATAA R: TCACATCCTCCGAATTCTTCTTC | 荧光定量引物 Fluorescent quantitative PCR primer |
Table 1 Primers for gene cloning and specific expressions
| 引物Primer | 序列Sequence(5'-3') | 用途Purpose |
|---|---|---|
| LbIRT2 | F: ATGGCTGCTCAAACCCAAGT R: TCACAACTCCATGCCAGTAGTAGA | 基因克隆 Gene clone |
| LbFRO | F: ATGGGTGAACTCTCAGGGCAA R: CTAGAGGTCAAAATTGTGGCAGTTG | 基因克隆 Gene clone |
| LbNRAMP3 | F: ATGCCTCCACACGATGAAGAACA R: TCAACTCTCTATGCTCGTGATACTCTT | 基因克隆 Gene clone |
| LbActin | F: TCTACGAGGGTTACGCTTTG R: TCCCGTTCAGCAGTGGTT | 枸杞内参引物 Internal primer of Lycium barbarum L. |
| qRT-LbFRO | F: TCAAGCCACCCAGGGAACT R: CATGAAAATTGGTAAACTGAACATCA | 荧光定量引物 Fluorescent quantitative PCR primer |
| qRT-LbNRAMP3 | F: GCAGAGAGCGTTAATAACAAGAAGTTG R: TTAAGCCACTCGTTGAGAACATCT | 荧光定量引物 Fluorescent quantitative PCR primer |
| qRT-LbIRT1 | F: AGGAGCAATTCATCAAGATGCA R: TGCTACATGCTCCGGCTACTAG | 荧光定量引物 Fluorescent quantitative PCR primer |
| qRT-LbIRT2 | F: TCCAAATTACCATTCCTAAAGACCAT R: CGAACCCACTGTGATCTTTGAG | 荧光定量引物 Fluorescent quantitative PCR primer |
| qRT-LbYSL | F: TGCAGTGTTCATGAGCGGTATAA R: TCACATCCTCCGAATTCTTCTTC | 荧光定量引物 Fluorescent quantitative PCR primer |

Fig. 1 Screening and hot map of ion transporter genes in the leaf tissue and root tissue of L. barbarum T12hL/CKL: The genes changed in the leaves at the 12th hour of Cd stress with the transcriptome data of the control; T48hL/CKL: the genes changed in the leaves at the 48th hour of Cd stress with the transcriptome data of the control; T12hR/CKR: the genes changed in the roots at the 12th hour of Cd stress with the transcriptome data of the control; T48hR/CKR: the genes changed in the roots at the 48th hour of Cd stress, the transcriptome data of the control
| 基因ID Gene ID | 基因名称 Gene name | log2 (T12hR/CKR) | log2 (T48hR/CKR) | 长度 Length/bp | 相似度 Similarity/% | 筛选部位 Isolation location |
|---|---|---|---|---|---|---|
| CL7788.Contig3 | 离子转运蛋白基因 Ion transport protein | 6.67 | 6.82 | 1 948 | 89.94 | 根系Root |
| Unigene39380 | OPT寡肽转运蛋白基因 OPT oligopeptide transporter | 8.372 403 | 10.0 693 | 1 739 | 99.47 | 根系Root |
| Unigene61286 | 锌转运体5 Zinc transporter 5-like | 4.88 | 5.86 | 1 431 | 82.17 | 根系Root |
| CL11868.Contig2 | ZIP家族锌转运蛋白基因 ZIP Zinc transporter | 7.28 | 7.23 | 1 167 | 98.77 | 根系Root |
| CL4044 | Nramp3金属离子转运蛋白基因 Metal transporter Nramp3-like | 5.3 | 4.23 493 | 3 830 | 92.62 | 根系Root |
| CL1976.Contig4_ | 机械敏感通道蛋白基因8 Mechanosensitive ion channel protein 8-like | 7.852 761 | 7.92 | 3 088 | 72.81 | 根系Root |
| Unigene60599 | 黄色条纹蛋白基因YSL2 Metal-nicotianamine transporter YSL2-like | 2.178 219 | 2.525 766 | 2 161 | 86.78 | 根系Root |
| CL1155.Contig | 黄色条纹蛋白基因YSL3 Metal-nicotianamine transporter YSL3-like | 9.070 304 | 6.348 448 | 3 045 | 91.95 | 根系Root |
| CL3488.Contig3 | 铁氧还原酶基因6 Ferric reduction oxidase 6-like | 1.875 425 | 2.782 025 | 3 193 | 89.01 | 叶片Leaf |
| Unigene44038 | 铁氧还原酶基因8 Ferric reduction oxidase 8 | 2.515 402 | 2.019 701 | 1 935 | 89.08 | 叶片Leaf |
| CL9188.Contig6 | 液泡膜铁离子转运蛋白基因4 Vacuolar iron transporter homolog 4-like | 3.63 367 | 2.881 373 | 588 | 84.66 | 叶片Leaf |
| CL12735.Contig1 | 铁氧还原酶基因2 Ferric reduction oxidase 2-like | 3.527 493 | 4.06 572 | 2 158 | 91.53 | 叶片Leaf |
| Unigene21912 | 寡肽转运蛋白基因4 Oligopeptide transporter 4 | 2.264 557 | 0.763 512 | 1 713 | 98.03 | 叶片Leaf |
| CL1352.Contig23 | 多向耐药性PDR跨膜转运蛋白基因1 Pleiotropic drug resistance protein 1-like | 9.955 166 | 10.28 298 | 5 054 | 90.33 | 叶片Leaf |
| CL17567.Contig5 | Nramp6金属离子转运蛋白基因 Metal transporter Nramp6 | 6.758 793 | 6.640 834 | 1 159 | 95.06 | 叶片Leaf |
| Unigene36942 | Nramp5金属离子转运蛋白基因 Metal transporter Nramp5 | 8.152 608 | 7.285 907 | 415 | 95.48 | 叶片Leaf |
Table 2 Genes related to ion transporter selected from the root and leaf
| 基因ID Gene ID | 基因名称 Gene name | log2 (T12hR/CKR) | log2 (T48hR/CKR) | 长度 Length/bp | 相似度 Similarity/% | 筛选部位 Isolation location |
|---|---|---|---|---|---|---|
| CL7788.Contig3 | 离子转运蛋白基因 Ion transport protein | 6.67 | 6.82 | 1 948 | 89.94 | 根系Root |
| Unigene39380 | OPT寡肽转运蛋白基因 OPT oligopeptide transporter | 8.372 403 | 10.0 693 | 1 739 | 99.47 | 根系Root |
| Unigene61286 | 锌转运体5 Zinc transporter 5-like | 4.88 | 5.86 | 1 431 | 82.17 | 根系Root |
| CL11868.Contig2 | ZIP家族锌转运蛋白基因 ZIP Zinc transporter | 7.28 | 7.23 | 1 167 | 98.77 | 根系Root |
| CL4044 | Nramp3金属离子转运蛋白基因 Metal transporter Nramp3-like | 5.3 | 4.23 493 | 3 830 | 92.62 | 根系Root |
| CL1976.Contig4_ | 机械敏感通道蛋白基因8 Mechanosensitive ion channel protein 8-like | 7.852 761 | 7.92 | 3 088 | 72.81 | 根系Root |
| Unigene60599 | 黄色条纹蛋白基因YSL2 Metal-nicotianamine transporter YSL2-like | 2.178 219 | 2.525 766 | 2 161 | 86.78 | 根系Root |
| CL1155.Contig | 黄色条纹蛋白基因YSL3 Metal-nicotianamine transporter YSL3-like | 9.070 304 | 6.348 448 | 3 045 | 91.95 | 根系Root |
| CL3488.Contig3 | 铁氧还原酶基因6 Ferric reduction oxidase 6-like | 1.875 425 | 2.782 025 | 3 193 | 89.01 | 叶片Leaf |
| Unigene44038 | 铁氧还原酶基因8 Ferric reduction oxidase 8 | 2.515 402 | 2.019 701 | 1 935 | 89.08 | 叶片Leaf |
| CL9188.Contig6 | 液泡膜铁离子转运蛋白基因4 Vacuolar iron transporter homolog 4-like | 3.63 367 | 2.881 373 | 588 | 84.66 | 叶片Leaf |
| CL12735.Contig1 | 铁氧还原酶基因2 Ferric reduction oxidase 2-like | 3.527 493 | 4.06 572 | 2 158 | 91.53 | 叶片Leaf |
| Unigene21912 | 寡肽转运蛋白基因4 Oligopeptide transporter 4 | 2.264 557 | 0.763 512 | 1 713 | 98.03 | 叶片Leaf |
| CL1352.Contig23 | 多向耐药性PDR跨膜转运蛋白基因1 Pleiotropic drug resistance protein 1-like | 9.955 166 | 10.28 298 | 5 054 | 90.33 | 叶片Leaf |
| CL17567.Contig5 | Nramp6金属离子转运蛋白基因 Metal transporter Nramp6 | 6.758 793 | 6.640 834 | 1 159 | 95.06 | 叶片Leaf |
| Unigene36942 | Nramp5金属离子转运蛋白基因 Metal transporter Nramp5 | 8.152 608 | 7.285 907 | 415 | 95.48 | 叶片Leaf |
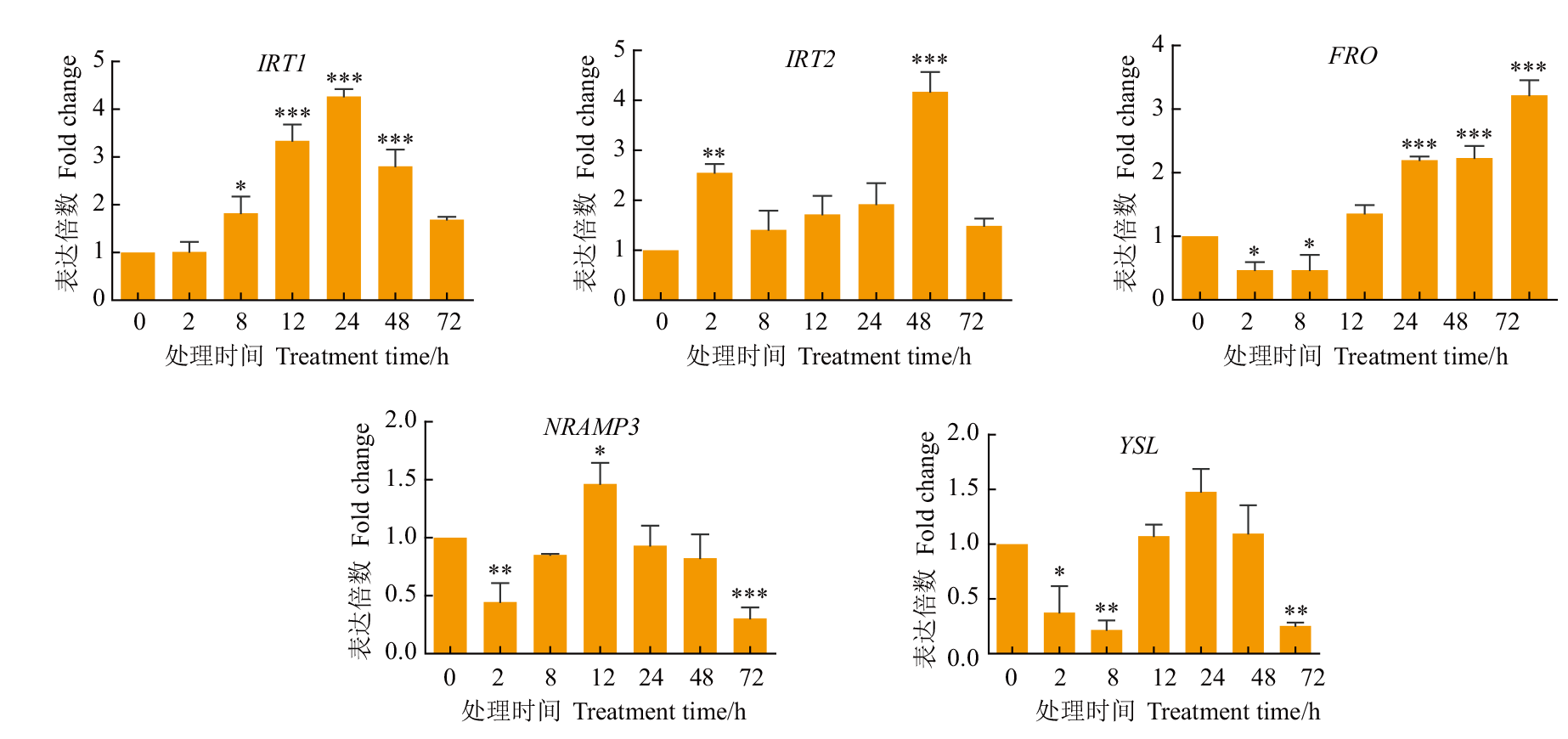
Fig. 2 Expression pattern of target gene under 200 μmol/L Fe-EDTA stress *, ** and *** denote P<0.05, P<0.01 and P<0.001, compared with the control(CK). The same below
| 基因名称 Gene name | 氨基酸长度 Amino acid length/aa | 分子量 Molecular weight/kD | 等电点 pI | 脂肪指数 Aliphatic index | 不稳定指数 Instability index | 亲疏水性 GRAVY |
|---|---|---|---|---|---|---|
| LbIRT2 | 215 | 22 607.41 | 8.81 | 97.26 | 40.50 | 0.340 |
| LbFRO | 740 | 83 469.16 | 8.48 | 112.19 | 36.60 | 0.371 |
| LbNRAMP3 | 511 | 56 347.21 | 5.83 | 119.10 | 37.17 | 0.556 |
Table 3 Physicochemical properties of functional genes responsible to iron and cadmium stress in L. barbarum
| 基因名称 Gene name | 氨基酸长度 Amino acid length/aa | 分子量 Molecular weight/kD | 等电点 pI | 脂肪指数 Aliphatic index | 不稳定指数 Instability index | 亲疏水性 GRAVY |
|---|---|---|---|---|---|---|
| LbIRT2 | 215 | 22 607.41 | 8.81 | 97.26 | 40.50 | 0.340 |
| LbFRO | 740 | 83 469.16 | 8.48 | 112.19 | 36.60 | 0.371 |
| LbNRAMP3 | 511 | 56 347.21 | 5.83 | 119.10 | 37.17 | 0.556 |
| 基因名称 Gene name | α-螺旋 α-helix/% | 延伸链 Extended strand/% | β-转角 β-turn/% | 无规则卷曲 Random coil/% |
|---|---|---|---|---|
| LbIRT2 | 55.35 | 12.09 | 8.37 | 24.19 |
| LbFRO | 37.03 | 21.35 | 3.11 | 38.51 |
| LbNRAMP3 | 55.58 | 12.33 | 3.91 | 28.18 |
Table 4 Prediction of secondary structure of functional genes in L. barbarum response to iron and cadmium stress
| 基因名称 Gene name | α-螺旋 α-helix/% | 延伸链 Extended strand/% | β-转角 β-turn/% | 无规则卷曲 Random coil/% |
|---|---|---|---|---|
| LbIRT2 | 55.35 | 12.09 | 8.37 | 24.19 |
| LbFRO | 37.03 | 21.35 | 3.11 | 38.51 |
| LbNRAMP3 | 55.58 | 12.33 | 3.91 | 28.18 |
| [1] | 王益民, 张宝琳. 我国枸杞属物种资源及发展对策[J]. 世界林业研究, 2021, 34(3): 107-111. |
| Wang YM, Zhang BL. Species resources of Lycium L. in China and their development strategies[J]. World For Res, 2021, 34(3): 107-111. | |
| [2] | 马鹏生, 朱溶月, 白长财, 等. 宁夏枸杞植物资源及产业发展调查[J]. 中成药, 2021, 43(11): 3245-3248. |
| Ma PS, Zhu RY, Bai CC, et al. Investigation on plant resources and industrial development of Lycium barbarum in Ningxia[J]. Chin Tradit Pat Med, 2021, 43(11): 3245-3248. | |
| [3] |
Buesseler KO, Andrews JE, Pike SM, et al. The effects of iron fertilization on carbon sequestration in the Southern Ocean[J]. Science, 2004, 304(5669): 414-417.
pmid: 15087543 |
| [4] |
Halliwell B, Gutteridge JMC. Biologically relevant metal ion-dependent hydroxyl radical generation An update[J]. FEBS Lett, 1992, 307(1): 108-112.
doi: 10.1016/0014-5793(92)80911-y pmid: 1322323 |
| [5] | 李云翔, 柯英, 罗健航, 等. 宁夏主要枸杞产地土壤环境质量现状与评价[J]. 中国土壤与肥料, 2016(2): 21-26. |
| Li YX, Ke Y, Luo JH, et al. Evaluation and present situation of soil environmental quality in the major wolfberry production areas of Ningxia[J]. Soil Fertil Sci China, 2016(2): 21-26. | |
| [6] |
王金龙, 王薇薇, 万平, 等. 缺铁胁迫对小豆幼苗生理特性的影响[J]. 生物技术通报, 2016, 32(10): 141-147.
doi: 10.13560/j.cnki.biotech.bull.1985.2016.10.017 |
| Wang JL, Wang WW, Wan P, et al. Effect of iron deficiency on physiological characteristics in seedlings of adzuki beans(Vigna angularis)[J]. Biotechnol Bull, 2016, 32(10): 141-147. | |
| [7] |
Nakanishi H, Ogawa I, Ishimaru Y, et al. Iron deficiency enhances cadmium uptake and translocation mediated by the Fe2+ transporters OsIRT1 and OsIRT2 in rice[J]. Soil Sci Plant Nutr, 2006, 52(4): 464-469.
doi: 10.1111/j.1747-0765.2006.00055.x URL |
| [8] | 吴非, 刘飞, 张苹, 等. 茄子和托鲁巴姆ZIP5和ZIP11基因对锌/镉毒害的响应分析[J]. 山西农业大学学报: 自然科学版, 2022, 42(2): 37-44. |
| Wu F, Liu F, Zhang P, et al. Response analyses of ZIP5 and ZIP11 genes in Solanum melongena and Solanum torvum to zinc/cadmium toxicity[J]. J Shanxi Agric Univ Nat Sci Ed, 2022, 42(2): 37-44. | |
| [9] | 简敏菲, 杨叶萍, 余厚平, 等. 不同浓度Cd2+胁迫对苎麻叶绿素及其光合荧光特性的影响[J]. 植物生理学报, 2015, 51(8): 1331-1338. |
| Jian MF, Yang YP, Yu HP, et al. Influences of different cadmium concentration stress on chlorophyll and its photosynthetic fluorescence characteristics of ramie(Boehmeria nivea)[J]. Plant Physiol J, 2015, 51(8): 1331-1338. | |
| [10] | 张林琳, 刘星星, 祝亚昕, 等. 机理I植物铁营养的吸收转运及信号调控机制研究进展[J]. 植物营养与肥料学报, 2021, 27(7): 1258-1272. |
| Zhang LL, Liu XX, Zhu YX, et al. Research progresses on iron uptake, translocation and their signaling pathways in strategy I plants[J]. J Plant Nutr Fertil, 2021, 27(7): 1258-1272. | |
| [11] | 李俊成, 于慧, 杨素欣, 等. 植物对铁元素吸收的分子调控机制研究进展[J]. 植物生理学报, 2016, 52(6): 835-842. |
| Li JC, Yu H, Yang SX, et al. Research progress of molecular regulation of iron uptake in plants[J]. Plant Physiol J, 2016, 52(6): 835-842. | |
| [12] |
Vasconcelos MW, Clemente TE, Grusak MA. Evaluation of constitutive iron reductase(AtFRO2)expression on mineral accumulation and distribution in soybean(Glycine max. L)[J]. Front Plant Sci, 2014, 5: 112.
doi: 10.3389/fpls.2014.00112 pmid: 24765096 |
| [13] |
Bernal M, Casero D, Singh V, et al. Transcriptome sequencing identifies SPL7-regulated copper acquisition genes FRO4/FRO5 and the copper dependence of iron homeostasis in Arabidopsis[J]. Plant Cell, 2012, 24(2): 738-761.
doi: 10.1105/tpc.111.090431 URL |
| [14] |
Mukherjee I, Campbell NH, Ash JS, et al. Expression profiling of the Arabidopsis ferric chelate reductase(FRO)gene family reveals differential regulation by iron and copper[J]. Planta, 2006, 223(6): 1178-1190.
doi: 10.1007/s00425-005-0165-0 pmid: 16362328 |
| [15] |
López-Millán AF, Duy D, Philippar K. Chloroplast iron transport proteins-function and impact on plant physiology[J]. Front Plant Sci, 2016, 7: 178.
doi: 10.3389/fpls.2016.00178 pmid: 27014281 |
| [16] |
Jain A, Connolly EL. Mitochondrial iron transport and homeostasis in plants[J]. Front Plant Sci, 2013, 4: 348.
doi: 10.3389/fpls.2013.00348 pmid: 24046773 |
| [17] |
Waters BM, Blevins DG, Eide DJ. Characterization of FRO1, a pea ferric-chelate reductase involved in root iron acquisition[J]. Plant Physiol, 2002, 129(1): 85-94.
doi: 10.1104/pp.010829 pmid: 12011340 |
| [18] |
Ling HQ, Bauer P, Bereczky Z, et al. The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots[J]. Proc Natl Acad Sci USA, 2002, 99(21): 13938-13943.
doi: 10.1073/pnas.212448699 URL |
| [19] | 贺晓燕, 龚义勤, 徐良, 等. 萝卜铁转运蛋白基因RsIRT1分子特征分析[J]. 南京农业大学学报, 2013, 36(6): 13-18. |
| He XY, Gong YQ, Xu L, et al. Molecular characterization of iron-regulated transporter gene RsIRT1 in radish[J]. J Nanjing Agric Univ, 2013, 36(6): 13-18. | |
| [20] | 谢昶琰, 金昕, 李岩, 等. 缺铁胁迫对杜梨幼苗生理及铁吸收和转运相关基因表达的影响[J]. 南京农业大学学报, 2019, 42(3): 465-473. |
| Xie CY, Jin X, Li Y, et al. Effects of iron deficiency stress on physiology and gene expression of iron absorption and transpotation in Pyrus betulaefolia[J]. J Nanjing Agric Univ, 2019, 42(3): 465-473. | |
| [21] |
Vatansever R, Filiz E, Ozyigit II. In silico analysis of Mn transporters(NRAMP1)in various plant species[J]. Mol Biol Rep, 2016, 43(3): 151-163.
doi: 10.1007/s11033-016-3950-x pmid: 26878855 |
| [22] |
Wu HL, Chen CL, Du J, et al. Co-overexpression FIT with Atb-HLH38 or AtbHLH39 in Arabidopsis-enhanced cadmium tolerance via increased cadmium sequestration in roots and improved iron homeostasis of shoots[J]. Plant Physiol, 2012, 158(2): 790-800.
doi: 10.1104/pp.111.190983 URL |
| [23] | 梁昕昕, 辛亚平, 贾雪, 等. 宁夏枸杞幼苗叶片中抗氧化酶及游离氨基酸对镉胁迫的响应[J]. 生态毒理学报, 2021, 16(6): 222-233. |
| Liang XX, Xin YP, Jia X, et al. Response of antioxidative enzymes activities and amino acids concentrations in leaf tissues of Lycium barbarum L. seedlings under cadmium stress[J]. Asian J Ecotoxicol, 2021, 16(6): 222-233. | |
| [24] |
Sun HP, Li F, Ruan QM, et al. Identification and validation of reference genes for quantitative real-time PCR studies in Hedera helix L[J]. Plant Physiol Biochem, 2016, 108: 286-294.
doi: 10.1016/j.plaphy.2016.07.022 URL |
| [25] |
乔孟欣, 李素贞, 陈景堂. 植物铁还原酶基因FRO的研究进展[J]. 生物技术通报, 2019, 35(7): 162-171.
doi: 10.13560/j.cnki.biotech.bull.1985.2018-1012 |
| Qiao MX, Li SZ, Chen JT. Research progress of Fe3+ reductase genes(FRO)in plants[J]. Biotechnol Bull, 2019, 35(7): 162-171. | |
| [26] |
Satbhai SB, Setzer C, Freynschlag F, et al. Natural allelic variation of FRO2 modulates Arabidopsis root growth under iron deficiency[J]. Nat Commun, 2017, 8: 15603.
doi: 10.1038/ncomms15603 URL |
| [27] |
Li LH, Cheng XD, Ling HQ. Isolation and characterization of Fe(III)-chelate reductase gene LeFRO1 in tomato[J]. Plant Mol Biol, 2004, 54(1): 125-136.
doi: 10.1023/B:PLAN.0000028774.82782.16 URL |
| [28] | 周正一. 苹果属小金海棠铁还原酶基因MxFRO4、MxFRO6的克隆与功能分析[D]. 哈尔滨: 东北农业大学, 2021. |
| Zhou ZY. Isolation and functional analysis of ferric reductase gene MxFRO4 and MxFRO6 in Malus xiaojinensis[D]. Harbin:Northeast Agricultural University, 2021. | |
| [29] | Finatto T, de Oliveira AC, Chaparro C, et al. Abiotic stress and genome dynamics: specific genes and transposable elements response to iron excess in rice[J]. Rice(NY), 2015, 8: 13. |
| [30] |
Ishimaru Y, Suzuki M, Tsukamoto T, et al. Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+[J]. Plant J, 2006, 45(3): 335-346.
doi: 10.1111/j.1365-313X.2005.02624.x pmid: 16412081 |
| [31] | 陈侨月. 蓝莓ZIP转运子的克隆及生物学功能研究[D]. 金华: 浙江师范大学, 2019. |
| Chen QY. Cloning and functional analysis of ZIP transporters in blueberry[D]. Jinhua: Zhejiang Normal University, 2019. | |
| [32] |
Eide D, Broderius M, Fett J, et al. A novel iron-regulated metal transporter from plants identified by functional expression in yeast[J]. Proc Natl Acad Sci USA, 1996, 93(11): 5624-5628.
doi: 10.1073/pnas.93.11.5624 pmid: 8643627 |
| [33] |
Vert G, Barberon M, Zelazny E, et al. Arabidopsis IRT2 cooperates with the high-affinity iron uptake system to maintain iron homeostasis in root epidermal cells[J]. Planta, 2009, 229(6): 1171-1179.
doi: 10.1007/s00425-009-0904-8 URL |
| [34] |
Bereczky Z, Wang HY, Schubert V, et al. Differential regulation of NRAMP and IRT metal transporter genes in wild type and iron uptake mutants of tomato[J]. J Biol Chem, 2003, 278(27): 24697-24704.
doi: 10.1074/jbc.M301365200 pmid: 12709425 |
| [35] | 王利华. 拟南芥AtNRAMP3转运蛋白的结构与功能研究[D]. 南京: 南京农业大学, 2017. |
| Wang LH. Structure and function of AtNRAMP3 transporter in Arabidopsis thaliana[D]. Nanjing: Nanjing Agricultural University, 2017. | |
| [36] | 陈邦兰, 龙涛, 高永峰, 等. 烟草NtNRAMP3.1基因克隆、表达及Cd转运特性分析[J]. 西南农业学报, 2022, 35(2): 438-445. |
| Chen BL, Long T, Gao YF, et al. Cloning, expression and Cd transport characteristic analysis of NtNRAMP3.1 gene from Nicotiana tabacum[J]. Southwest China J Agric Sci, 2022, 35(2): 438-445. | |
| [37] |
Thomine S, Wang R, Ward JM, et al. Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes[J]. Proc Natl Acad Sci USA, 2000, 97(9): 4991-4996.
doi: 10.1073/pnas.97.9.4991 pmid: 10781110 |
| [38] |
Pottier M, Oomen R, Picco C, et al. Identification of mutations allowing Natural Resistance Associated Macrophage Proteins(NRAMP)to discriminate against cadmium[J]. Plant J, 2015, 83(4): 625-637.
doi: 10.1111/tpj.12914 URL |
| [1] | LIU Hui, LU Yang, YE Xi-miao, ZHOU Shuai, LI Jun, TANG Jian-bo, CHEN En-fa. Comparative Transcriptome Analysis of Cadmium Stress Response Induced by Exogenous Sulfur in Tartary Buckwheat [J]. Biotechnology Bulletin, 2023, 39(5): 177-191. |
| [2] | JIANG Nan, SHI Yang, ZHAO Zhi-hui, LI Bin, ZHAO Yi-hui, YANG Jun-biao, YAN Jia-ming, JIN Yu-fan, CHEN Ji, HUANG Jin. Expression and Functional Analysis of OsPT1 Gene in Rice Under Cadmium Stress [J]. Biotechnology Bulletin, 2023, 39(1): 166-174. |
| [3] | HU Yan-jiao, CHEN Mei-feng, QIANG Yu, LI Hai-yan, LIU Jing, QIN Fan-xin. Alleviation Mechanisms of Zinc-selenium Interaction on the Cadmium Toxicity in Rice Under Cadmium Stress [J]. Biotechnology Bulletin, 2022, 38(4): 143-152. |
| [4] | ZU Guo-qiang, HU Zhe, WANG Qi, LI Guang-zhe, HAO Lin. Regulatory Role of Burkholderia sp. GD17 in Rice Seedling’s Responses to Cadmium Stress [J]. Biotechnology Bulletin, 2022, 38(4): 153-162. |
| [5] | YANG Fu-rong, WANG Xiao-hong, XIAO Qi, FANG Juan, LI Li-hua. Physiological Response of Hibiscus syriacus Varieties to Cadmium Stress and Evaluation of Cadmium Tolerance [J]. Biotechnology Bulletin, 2022, 38(1): 98-107. |
| [6] | WANG Zhu-cheng, LIU Hui, LI Rong-hua. Effects of Exogenous Sulfur on Photosynthetic Characteristics and Mineral Elements Absorption in Portulaca oleracea Under Cadmium Stress [J]. Biotechnology Bulletin, 2020, 36(3): 133-140. |
| [7] | WANG Zhu-cheng, LIU Hui, LI Rong-hua, CHEN Xin, LI Xin, LU Zhi-yuan. Physiological Mechanism of Exogenous Ethylene and Sulfur in Alleviating Cadmium Stress in Portulaca oleracea [J]. Biotechnology Bulletin, 2019, 35(10): 71-79. |
| [8] | MA Xiao-li, JI Rui-ping, TIAN Bao-hua, HUO Jian-xin. Effects of Nitric Oxide on Oxidative Damage Metabolism in Wheat Seedling Under Cadmium Stress [J]. Biotechnology Bulletin, 2017, 33(5): 102-107. |
| [9] | HE Ting-ting, SONG Ting, WANG Chao, ZHANG Chang-bin, WANG Hai-yan. Screening of Reference Genes in Bacillus pumilus by Real-time Fluorescence Quantitative PCR [J]. Biotechnology Bulletin, 2016, 32(11): 99-106. |
| [10] | Wang Fengjun, Liu Shasha, Feng Junli. Rapid Identification of Four Kinds of Decay Fruit Moths by Real-time Fluorescence Quantitative PCR [J]. Biotechnology Bulletin, 2015, 31(2): 78-83. |
| [11] | Wei Di, Gao Jing, Chi Zhenfen, Zhang Guirong, Nie Lingyun. A Novel Method for Detecting Activity of Mitochondria-targeted Nuclease [J]. Biotechnology Bulletin, 2015, 31(12): 81-90. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||