








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (9): 176-182.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0275
Previous Articles Next Articles
WANG Teng-hui1( ), GE Wen-dong1, LUO Ya-fang1, FAN Zhen-yu1, WANG Yu-shu1,2(
), GE Wen-dong1, LUO Ya-fang1, FAN Zhen-yu1, WANG Yu-shu1,2( )
)
Received:2023-03-24
Online:2023-09-26
Published:2023-10-24
Contact:
WANG Yu-shu
E-mail:372838418@qq.com;wangys1019@126.com
WANG Teng-hui, GE Wen-dong, LUO Ya-fang, FAN Zhen-yu, WANG Yu-shu. Gene Mapping of Kale White Leaves Based on Whole Genome Re-sequencing of Extreme Mixed Pool(BSA)[J]. Biotechnology Bulletin, 2023, 39(9): 176-182.
| 群体 Group | 总株数 Total plants | 红叶株数 No. of red-leaf plants | 白叶株数 No. of white-leaf plants | 红叶∶白叶 Red leaf∶White leaf | 卡方值 χ2 | 检验值 χ20.05 |
|---|---|---|---|---|---|---|
| WR | 20 | 20 | 0 | - | - | - |
| WB | 20 | 0 | 20 | - | - | - |
| F1 | 20 | 20 | 0 | - | - | - |
| F2 | 1 300 | 982 | 318 | 3.09∶1 | 0.20 | 3.84 |
Table 1 Segregation of the crosses between ‘WR’ and ‘WB’ in progeny populations
| 群体 Group | 总株数 Total plants | 红叶株数 No. of red-leaf plants | 白叶株数 No. of white-leaf plants | 红叶∶白叶 Red leaf∶White leaf | 卡方值 χ2 | 检验值 χ20.05 |
|---|---|---|---|---|---|---|
| WR | 20 | 20 | 0 | - | - | - |
| WB | 20 | 0 | 20 | - | - | - |
| F1 | 20 | 20 | 0 | - | - | - |
| F2 | 1 300 | 982 | 318 | 3.09∶1 | 0.20 | 3.84 |
| 样品 Sample | Clean reads数 Number of clean reads | 比对reads数 Mapped reads | 参考基因组比对 Map ratio/% | 平均覆盖深度 Average cover depth | 基因组覆盖度 Coverage/% | 碱基数质量值大于Q20/% |
|---|---|---|---|---|---|---|
| W12 | 122 570 282 | 117 542 515 | 95.90 | 29.03 | 88.05 | 98.35 |
| W06 | 156 059 918 | 148 403 662 | 95.09 | 35.68 | 88.38 | 98.17 |
| L-bulk | 370 780 056 | 352 633 985 | 95.11 | 81.94 | 91.05 | 98.19 |
| B-bulk | 324 884 700 | 307 268 196 | 94.58 | 73.98 | 91.12 | 97.81 |
Table 2 Sequencing comparison results of different samples
| 样品 Sample | Clean reads数 Number of clean reads | 比对reads数 Mapped reads | 参考基因组比对 Map ratio/% | 平均覆盖深度 Average cover depth | 基因组覆盖度 Coverage/% | 碱基数质量值大于Q20/% |
|---|---|---|---|---|---|---|
| W12 | 122 570 282 | 117 542 515 | 95.90 | 29.03 | 88.05 | 98.35 |
| W06 | 156 059 918 | 148 403 662 | 95.09 | 35.68 | 88.38 | 98.17 |
| L-bulk | 370 780 056 | 352 633 985 | 95.11 | 81.94 | 91.05 | 98.19 |
| B-bulk | 324 884 700 | 307 268 196 | 94.58 | 73.98 | 91.12 | 97.81 |
| 变异位点信息 Variation sites information | WR | WB | R-bulk | B-bulk |
|---|---|---|---|---|
| 基因区间 Intergenic region | 525 758 | 529 072 | 555 294 | 555 413 |
| 非同义突变 Non-synonymous | 266 962 | 270 929 | 283 962 | 283 910 |
| 同义突变 Synonymous | 150 618 | 152 757 | 159 396 | 159 535 |
| 起始密码子丢失 Start lost | 611 | 628 | 672 | 668 |
| 终止密码子丢失 Stop_lost | 6 645 | 6 703 | 7 028 | 7 075 |
| 终止密码子获得 Stop_Gained | 13 829 | 13 994 | 14 600 | 14 596 |
| 5'非翻译区 UTR_5_Prime | 489 | 484 | 511 | 512 |
| SNP总数 SNP total | 964 912 | 979 644 | 1 021 463 | 1 021 699 |
Table 3 Annotation of variation sites
| 变异位点信息 Variation sites information | WR | WB | R-bulk | B-bulk |
|---|---|---|---|---|
| 基因区间 Intergenic region | 525 758 | 529 072 | 555 294 | 555 413 |
| 非同义突变 Non-synonymous | 266 962 | 270 929 | 283 962 | 283 910 |
| 同义突变 Synonymous | 150 618 | 152 757 | 159 396 | 159 535 |
| 起始密码子丢失 Start lost | 611 | 628 | 672 | 668 |
| 终止密码子丢失 Stop_lost | 6 645 | 6 703 | 7 028 | 7 075 |
| 终止密码子获得 Stop_Gained | 13 829 | 13 994 | 14 600 | 14 596 |
| 5'非翻译区 UTR_5_Prime | 489 | 484 | 511 | 512 |
| SNP总数 SNP total | 964 912 | 979 644 | 1 021 463 | 1 021 699 |
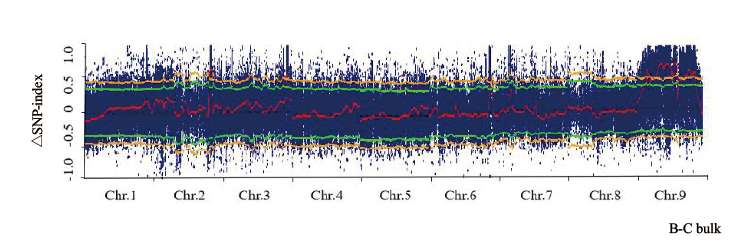
Fig. 1 △SNP-index map of all chromosomes The red curve represents the Delta SNP-index value after the sliding window, the yellow curve represents the 99% confidence interva
| 染色体 Chromosome | 起始位置 Start/bp | 终止位置 End/bp | 关联区域大小 Size/Mb | 基因数量 No. of genes |
|---|---|---|---|---|
| Chr.2 | 35 100 000 | 37 050 000 | 3.95 | 20 |
| Chr.3 | 51 600 000 | 52 800 000 | 1.20 | 38 |
| Chr.9 | 5 850 000 | 7 850 000 | 2.00 | 20 |
| Chr.9 | 9 150 000 | 10 600 000 | 1.45 | 14 |
| Chr.9 | 11 900 000 | 21 650 000 | 9.75 | 199 |
| Chr.9 | 22 000 000 | 24 250 000 | 2.25 | 16 |
| Chr.9 | 28 100 000 | 33 050 000 | 4.95 | 151 |
| Chr.9 | 33 250 000 | 35 350 000 | 2.10 | 53 |
| 总计Total | — | — | 27.65 | 510 |
Table 4 Correlation regions obtained by SNP correlation analysis methods
| 染色体 Chromosome | 起始位置 Start/bp | 终止位置 End/bp | 关联区域大小 Size/Mb | 基因数量 No. of genes |
|---|---|---|---|---|
| Chr.2 | 35 100 000 | 37 050 000 | 3.95 | 20 |
| Chr.3 | 51 600 000 | 52 800 000 | 1.20 | 38 |
| Chr.9 | 5 850 000 | 7 850 000 | 2.00 | 20 |
| Chr.9 | 9 150 000 | 10 600 000 | 1.45 | 14 |
| Chr.9 | 11 900 000 | 21 650 000 | 9.75 | 199 |
| Chr.9 | 22 000 000 | 24 250 000 | 2.25 | 16 |
| Chr.9 | 28 100 000 | 33 050 000 | 4.95 | 151 |
| Chr.9 | 33 250 000 | 35 350 000 | 2.10 | 53 |
| 总计Total | — | — | 27.65 | 510 |
| 基因ID Gene ID | 染色体位置 Chromosomal location/bp | 基因功能注释 Gene function annotation |
|---|---|---|
| Bol030253 | Chr.9:19341984-19344641 | 五肽重复(PPR)超家族蛋白Pentatricopeptide repeat(PPR)superfamily protein |
| Bol029431 | Chr.9:18884039-18885364 | 跨膜蛋白161AB蛋白Transmembrane protein 161AB protein |
| Bol012077 | Chr.9:12469942-12472086 | 跨膜蛋白,推测(DUF3537)Transmembrane protein, putative(DUF3537) |
| Bol007709 | Chr.9:14272763-14273749 | 跨膜蛋白Transmembrane protein |
| Bol030318 | Chr.9:20636604-20637310 | 跨膜蛋白Transmembrane protein |
| Bol030235 | Chr.9:19061948-19069105 | 胡萝卜素通过隐黄质转化为玉米黄质Carotene to zeaxanthin via cryptoxanthin |
| Bol030286 | Chr.9:19934118-19938365 | 编码一种定位于叶绿体和线粒体的FtsH蛋白酶Encodes an FtsH protease that is localized to the chloroplast and the mitochondrion |
| Bol005195 | Chr.9:14477658-14478362 | 叶绿体atp合酶δ亚基ATP synthase subunit delta, chloroplast |
Table 5 Functional annotation of candidate genes
| 基因ID Gene ID | 染色体位置 Chromosomal location/bp | 基因功能注释 Gene function annotation |
|---|---|---|
| Bol030253 | Chr.9:19341984-19344641 | 五肽重复(PPR)超家族蛋白Pentatricopeptide repeat(PPR)superfamily protein |
| Bol029431 | Chr.9:18884039-18885364 | 跨膜蛋白161AB蛋白Transmembrane protein 161AB protein |
| Bol012077 | Chr.9:12469942-12472086 | 跨膜蛋白,推测(DUF3537)Transmembrane protein, putative(DUF3537) |
| Bol007709 | Chr.9:14272763-14273749 | 跨膜蛋白Transmembrane protein |
| Bol030318 | Chr.9:20636604-20637310 | 跨膜蛋白Transmembrane protein |
| Bol030235 | Chr.9:19061948-19069105 | 胡萝卜素通过隐黄质转化为玉米黄质Carotene to zeaxanthin via cryptoxanthin |
| Bol030286 | Chr.9:19934118-19938365 | 编码一种定位于叶绿体和线粒体的FtsH蛋白酶Encodes an FtsH protease that is localized to the chloroplast and the mitochondrion |
| Bol005195 | Chr.9:14477658-14478362 | 叶绿体atp合酶δ亚基ATP synthase subunit delta, chloroplast |
| [1] | 孙日飞, 张淑江, 章时蕃, 等. 紫红色大白菜种质的创新研究[J]. 园艺学报, 2006, 33(5): 1032. |
| Sun RF, Zhang SJ, Zhang SF, et al. Research on creation of purple Chinese cabbage germplasm[J]. Acta Hortic Sin, 2006, 33(5): 1032. | |
| [2] |
Li HB, Zhu LX, Yuan GG, et al. Fine mapping and candidate gene analysis of an anthocyanin-rich gene, BnaA.PL1, conferring purple leaves in Brassica napus L.[J]. Mol Genet Genomics, 2016, 291(4): 1523-1534.
doi: 10.1007/s00438-016-1199-7 URL |
| [3] |
Guo N, Wu J, Zheng SN, et al. Anthocyanin profile characterization and quantitative trait locus mapping in zicaitai(Brassica rapa L. ssp. chinensis var. purpurea)[J]. Mol Breeding, 2015, 35(5): 113.
doi: 10.1007/s11032-015-0237-1 URL |
| [4] |
Zhu PF, Cheng MM, Feng X, et al. Mapping of Pi, a gene conferring pink leaf in ornamental kale(Brassica oleracea L. var. acephala DC)[J]. Euphytica, 2016, 207(2): 377-385.
doi: 10.1007/s10681-015-1555-4 URL |
| [5] |
He Q, Lu QQ, He YT, et al. Dynamic changes of the anthocyanin biosynthesis mechanism during the development of heading Chinese cabbage(Brassica rapa L.) and Arabidopsis under the control of BrMYB2[J]. Front Plant Sci, 2020, 11: 593766.
doi: 10.3389/fpls.2020.593766 URL |
| [6] | 叶沈华, 马晓伟, 杨杰, 等. 甘蓝型油菜叶片白化分子机理研究[J]. 植物遗传资源学报, 2023, 1: 1-18 |
| Ye SH, Ma XW, Yang J, et al. Molecular mechanisms of albino leaves in Brassica napus[J]. J Plant Gen Res, 2023, 1: 1-18. | |
| [7] |
Jiang XF, Zhao H, Guo F, et al. Transcriptomic analysis reveals mechanism of light-sensitive albinism in tea plant Camellia sinensis ‘Huangjinju ’[J]. BMC Plant Biol, 2020, 20(1): 216.
doi: 10.1186/s12870-020-02425-0 |
| [8] | 王容, 李博, 刘刚, 等. 大麦阶段性低温诱导白化突变体的转录组分析[J]. 麦类作物学报, 2023, 43(1): 64-74. |
| Wang R, Li B, Liu G, et al. Transcription analysis of stage specific low temperature induced albinism in barley[J]. J Triticeae Crops, 2023, 43(1): 64-74. | |
| [9] |
Lu HF, Lin T, Klein J, et al. QTL-seq identifies an early flowering QTL located near flowering locus T in cucumber[J]. Theor Appl Genet, 2014, 127(7): 1491-1499.
doi: 10.1007/s00122-014-2313-z URL |
| [10] |
Illa-Berenguer E, Van Houten J, Huang ZJ, et al. Rapid and reliable identification of tomato fruit weight and locule number loci by QTL-seq[J]. Theor Appl Genet, 2015, 128(7): 1329-1342.
doi: 10.1007/s00122-015-2509-x pmid: 25893466 |
| [11] |
张尧锋, 张冬青, 余华胜, 等. 基于极端混合池(BSA)全基因组重测序的甘蓝型油菜有限花序基因定位[J]. 中国农业科学, 2018, 51(16): 3029-3039.
doi: 10.3864/j.issn.0578-1752.2018.16.001 |
| Zhang YF, Zhang DQ, Yu HS, et al. Location and mapping of the determinate growth habit of Brassica napus by bulked segregant analysis(BSA)using whole genome re-sequencing[J]. Sci Agric Sin, 2018, 51(16): 3029-3039. | |
| [12] |
乔军, 刘婧, 李素文, 等. 基于极端混合池全基因组重测序的茄子萼下果色基因预测[J]. 园艺学报, 2022, 49(3): 613-621.
doi: 10.16420/j.issn.0513-353x.2021-0018 |
| Qiao J, Liu J, Li SW, et al. Prediction of fruit color genes under the Calyx of eggplant based on genome-wide resequencing in an extreme mixing pool[J]. Acta Hortic Sin, 2022, 49(3): 613-621. | |
| [13] |
严昕, 项超, 刘荣, 等. 基于BSA-seq技术对豌豆花色基因的精细定位[J]. 作物学报, 2023, 49(4): 1006-1015.
doi: 10.3724/SP.J.1006.2023.24055 |
| Yan X, Xiang C, Liu R, et al. Fine mapping of pea flower color genes based on BSA-seq technology[J]. Acta Agron Sin, 2023, 49(4): 1006-1015. | |
| [14] |
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data[J]. Bioinformatics, 2014, 30(15): 2114-2120.
doi: 10.1093/bioinformatics/btu170 pmid: 24695404 |
| [15] |
Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools[J]. Bioinformatics, 2009, 25(16): 2078-2079.
doi: 10.1093/bioinformatics/btp352 pmid: 19505943 |
| [16] |
Mckenna A, Hanna M, Banks E, et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data[J]. Genome Res, 2010, 20(9): 1297-1303.
doi: 10.1101/gr.107524.110 pmid: 20644199 |
| [17] |
Wang K, Li MY, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data[J]. Nucleic Acids Research, 2010, 38(16): e164.
doi: 10.1093/nar/gkq603 URL |
| [18] |
Takagi H, Abe A, Yoshida K, et al. QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations[J]. Plant J, 2013, 74(1): 174-183.
doi: 10.1111/tpj.2013.74.issue-1 URL |
| [19] |
Song B, Stöcklin J, Armbruster WS, et al. Reversible colour change in leaves enhances pollinator attraction and reproductive success in Saururus chinensis(Saururaceae)[J]. Ann bot, 2018, 121(4): 641-650.
doi: 10.1093/aob/mcx195 URL |
| [20] |
Sun JF, Gong YB, Renner SS, et al. Multifunctional bracts in the dove tree Davidia involucrata(Nyssaceae: Cornales): rain protection and pollinator attraction[J]. Am Nat, 2008, 171(1): 119-124.
doi: 10.1086/523953 URL |
| [21] |
Maiwald D, Dietzmann A, Jahns P, et al. Knock-out of the genes coding for the Rieske protein and the ATP-synthase delta-subunit of Arabidopsis. Effects on photosynthesis, thylakoid protein composition, and nuclear chloroplast gene expression[J]. Plant Physiol, 2003, 133(1): 191-202.
doi: 10.1104/pp.103.024190 URL |
| [22] |
Colcombet J, Lopez-Obando M, Heurtevin L, et al. Systematic study of subcellular localization of Arabidopsis PPR proteins confirms a massive targeting to organelles[J]. RNA Biol, 2013, 10(9): 1557-1575.
doi: 10.4161/rna.26128 pmid: 24037373 |
| [23] |
Liu XY, Jiang RC, Wang Y, et al. ZmPPR26, a DYW-type pentatricopeptide repeat protein, is required for C-to-U RNA editing at atpA-1148 in maize chloroplasts[J]. J Exp Bot, 2021, 72(13): 4809-4821.
doi: 10.1093/jxb/erab185 URL |
| [24] |
刘胜坤, 孙世磊, 董鲁朋, 等. 玉米黄叶基因ZmNPPR5的克隆[J]. 植物遗传资源学报, 2023, 24(2): 388-395.
doi: 10.13430/j.cnki.jpgr.20220918002 |
| Liu SK, Sun SL, Dong LP, et al. Cloning of a new yellow leaf gene ZmNPPR5 in maize[J]. J Plant Genet Resour, 2023, 24(2): 388-395. | |
| [25] |
Wagner R, von Sydow L, Aigner H, et al. Deletion of FtsH11 protease has impact on chloroplast structure and function in Arabidopsis thaliana when grown under continuous light[J]. Plant Cell Environ, 2016, 39(11): 2530-2544.
doi: 10.1111/pce.v39.11 URL |
| [26] |
Xia H, Zhou Y, Lin Z, et al. Characterization and functional validation of β-carotene hydroxylase AcBCH genes in Actinidia chinensis[J]. Hortic Res, 2022, 9: uhac063.
doi: 10.1093/hr/uhac063 URL |
| [27] |
Diretto G, Wwlsch R, Tavazza R et al. Silencing of beta-carotene hydroxylase increases total carotenoid and beta-carotene levels in potato tubers[J]. BMC Plant Biol, 2007, 7(1): 11.
doi: 10.1186/1471-2229-7-11 |
| [28] | 宋江华, 张立新. 植物跨膜蛋白研究进展[J]. 生物学杂志, 2009, 26(6): 62-64. |
| Song JH, Zhang LX. Progress on the transmembrane protein in plants[J]. J Biol, 2009, 26(6): 62-64. |
| [1] | WU Yuan-ming, LIN Jia-yi, LIU Yu-xi, LI Dan-ting, ZHANG Zong-qiong, ZHENG Xiao-ming, PANG Hong-bo. Identification of Rice Plant Height-associated QTL Using BSA-seq and RNA-seq [J]. Biotechnology Bulletin, 2023, 39(8): 173-184. |
| [2] | FANG Lan, LI Yan-yan, JIANG Jian-wei, CHENG Sheng, SUN Zheng-xiang, ZHOU Yi. Isolation, Identification and Growth-promoting Characteristics of Endohyphal Bacterium 7-2H from Endophytic Fungi of Spiranthes sinensis [J]. Biotechnology Bulletin, 2023, 39(8): 272-282. |
| [3] | GUO Shao-hua, MAO Hui-li, LIU Zheng-quan, FU Mei-yuan, ZHAO Ping-yuan, MA Wen-bo, LI Xu-dong, GUAN Jian-yi. Whole Genome Sequencing and Comparative Genome Analysis of a Fish-derived Pathogenic Aeromonas Hydrophila Strain XDMG [J]. Biotechnology Bulletin, 2023, 39(8): 291-306. |
| [4] | ZHANG Zhi-xia, LI Tian-pei, ZENG Hong, ZHU Xi-xian, YANG Tian-xiong, MA Si-nan, HUANG Lei. Genome Sequencing and Bioinformatics Analysis of Gelidibacter sp. PG-2 [J]. Biotechnology Bulletin, 2023, 39(3): 290-300. |
| [5] | HE Meng-ying, LIU Wen-bin, LIN Zhen-ming, LI Er-tong, WANG Jie, JIN Xiao-bao. Whole Genome Sequencing and Analysis of an Anti Gram-positive Bacterium Gordonia WA4-43 [J]. Biotechnology Bulletin, 2023, 39(2): 232-242. |
| [6] | ZHANG Ao-jie, LI Qing-yun, SONG Wen-hong, YAN Shao-hui, TANG Ai-xing, LIU You-yan. Whole Genome Sequencing Analysis of a Phenol-degrading Strain Alcaligenes faecalis JF101 [J]. Biotechnology Bulletin, 2023, 39(10): 292-303. |
| [7] | WANG Shuai, LV Hong-rui, ZHANG Hao, WU Zhan-wen, XIAO Cui-hong, SUN Dong-mei. Whole-Genome Sequencing Identification of Phosphate-solubilizing Bacteria PSB-R and Analysis of Its Phosphate-solubilizing Properties [J]. Biotechnology Bulletin, 2023, 39(1): 274-283. |
| [8] | WEN Chang, LIU Chen, LU Shi-yun, XU Zhong-bing, AI Chao-fan, LIAO Han-peng, ZHOU Shun-gui. Biological Characteristics and Genome Analysis of a Novel Multidrug-resistant Shigella flexneri Phage [J]. Biotechnology Bulletin, 2022, 38(9): 127-135. |
| [9] | LI Ji-hong, JING Yu-ling, MA Gui-zhen, GUO Rong-jun, LI Shi-dong. Genome Construction of Achromobacter 77 and Its Characteristics on Chemotaxis and Antibiotic Resistance [J]. Biotechnology Bulletin, 2022, 38(9): 136-146. |
| [10] | ZHOU Shi-chen, YI Zhi-ben, WANG Xin-yi, YANG Xiao-ying, SUN Li-na, LUAN Wei-jiang, LIANG Shan-shan. Genetic Analysis and Gene Mapping of Sorghum Double-grain Mutant Dgs [J]. Biotechnology Bulletin, 2022, 38(7): 171-177. |
| [11] | ZHANG Ze-ying, FAN Qing-feng, DENG Yun-feng, WEI Ting-zhou, ZHOU Zheng-fu, ZHOU Jian, WANG Jin, JIANG Shi-jie. Whole Genome Sequencing and Comparative Genomic Analysis of a High-yield Lipase-producing Strain WCO-9 [J]. Biotechnology Bulletin, 2022, 38(10): 216-225. |
| [12] | CHEN Ti-qiang, XU Xiao-lan, SHI Lin-chun, ZHONG Li-Yi. Sequencing and Analysis of the Whole Genome of Zizhi Cultivar ‘Wuzhi No.2’(Ganoderma sp. strain Zizhi S2) [J]. Biotechnology Bulletin, 2021, 37(11): 42-56. |
| [13] | GUO He-bao, WANG Xing, HE Shan-wen, ZHANG Xiao-xia. Phenotypic Characteristics Combined with Genomic Analysis to Identify Different Colony Morphology Bacillus velezensis ACCC 19742 [J]. Biotechnology Bulletin, 2020, 36(2): 142-148. |
| [14] | HAN Li-jie, CAI Hong-wei. Progress on Genetic Research of Sorghum Grain Weight [J]. Biotechnology Bulletin, 2019, 35(5): 15-27. |
| [15] | Dong Bin, Li Rongxi, Huang Yongfang, Hong Wenhong, Tan Sha,. Application of Molecular Markers in Studies of Camellia oleifera [J]. Biotechnology Bulletin, 2015, 31(6): 74-80. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||