








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (1): 186-193.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0336
Previous Articles Next Articles
LIU Xing-yu1( ), LI Jie2, ZHU Long-jiao3, LI Xiang-yang2, XU Wen-tao3(
), LI Jie2, ZHU Long-jiao3, LI Xiang-yang2, XU Wen-tao3( )
)
Received:2023-04-11
Online:2024-01-26
Published:2024-02-06
Contact:
XU Wen-tao
E-mail:liuxingyu0321@163.com;xuwentao@cau.edu.cn
LIU Xing-yu, LI Jie, ZHU Long-jiao, LI Xiang-yang, XU Wen-tao. Aptamer of Pseudomonas aeruginosa: Acquiring and Application[J]. Biotechnology Bulletin, 2024, 40(1): 186-193.
| 靶标 Target | 筛选方式SELEX type | 序列 Sequence(5'-3') | Kd/(nmol·L-1) | 亲和力检测方式 Affinity detection method | 长度Length/nt | 参考文献Reference |
|---|---|---|---|---|---|---|
| 全菌 | Cell-SELEX | CCCCCGTTGCTTTCGCTTTTCCTTTCGCTTGT-GTTCGTTTCGTCCCTGCTTCCTTCCTTCCTTC-CTTTCTTG | 17.27 ± 5.00 | 流式细胞术 | 72 | [ |
| 全菌 | Cell-SELEX | ATGCACTCTTCTATCGGTAGTTGAGGGTGCGG | 12.5±2.4 | 流式细胞术 | 32 | [ |
| 外毒素A | Decoy-SELEX | TGTACCGTCTGAGCGATTCGTACCATAGGGTGCTTTTCAAGGCCACACGTTAGTGTAAGCCAGTCAGTGTTAAGGAGTGC | 4 200-4 500 | 表面等离子体共振 | 80 | [ |
| 脂多糖 | Capture-SELEX | CCTTCTAACAGAATGTTGTTAGATAGC | 46.2±9.5 | 等热滴定量热 | 27 | [ |
| 全菌 | Cell-SELEX | GGGAGGACFGAUFGCFGGGAACFUFGAAGAGUFUFUFGAUFCFAUFGGCFUFCFAGAUFUFGAACFGCFUFGGCFGGCFAGACFGACFUCFGCFGCFGA | — | — | 73 | [ |
| 全菌 | WB-SELEX | TGGACCTTGCGATTGACAGCAGACATGAGTCAGGACGTGACCGCTGGACCTTGCGATTGACAGCAGACATGAGTCAGGAC | 15.16±3.62 | 流式细胞术 | 80 | [ |
| 全菌 | Cell-SELEX | CCCCCGTTGCTTTCGCTTTTCCTTTCGCTTTTGTTCGTTTCGTCCCTGCTTCCTTTCTTG | 14.55 | 地高辛标记适配体 | 60 | [ |
| 丁酰基-高丝氨酸 | Structure-switching SELEX | CCATCCACACTCCGCAAGTGGGGAGGGGAGAGACGACGATCCTGTGGGTTTTCTGCAGTGAGTCGTGTTTTCGACTTATTGCGTCGGCTGCCTCTACAT | 28.47 | 流式细胞术 | 99 | [ |
Table 1 SELEX of Pseudomonas aeruginosa aptamers
| 靶标 Target | 筛选方式SELEX type | 序列 Sequence(5'-3') | Kd/(nmol·L-1) | 亲和力检测方式 Affinity detection method | 长度Length/nt | 参考文献Reference |
|---|---|---|---|---|---|---|
| 全菌 | Cell-SELEX | CCCCCGTTGCTTTCGCTTTTCCTTTCGCTTGT-GTTCGTTTCGTCCCTGCTTCCTTCCTTCCTTC-CTTTCTTG | 17.27 ± 5.00 | 流式细胞术 | 72 | [ |
| 全菌 | Cell-SELEX | ATGCACTCTTCTATCGGTAGTTGAGGGTGCGG | 12.5±2.4 | 流式细胞术 | 32 | [ |
| 外毒素A | Decoy-SELEX | TGTACCGTCTGAGCGATTCGTACCATAGGGTGCTTTTCAAGGCCACACGTTAGTGTAAGCCAGTCAGTGTTAAGGAGTGC | 4 200-4 500 | 表面等离子体共振 | 80 | [ |
| 脂多糖 | Capture-SELEX | CCTTCTAACAGAATGTTGTTAGATAGC | 46.2±9.5 | 等热滴定量热 | 27 | [ |
| 全菌 | Cell-SELEX | GGGAGGACFGAUFGCFGGGAACFUFGAAGAGUFUFUFGAUFCFAUFGGCFUFCFAGAUFUFGAACFGCFUFGGCFGGCFAGACFGACFUCFGCFGCFGA | — | — | 73 | [ |
| 全菌 | WB-SELEX | TGGACCTTGCGATTGACAGCAGACATGAGTCAGGACGTGACCGCTGGACCTTGCGATTGACAGCAGACATGAGTCAGGAC | 15.16±3.62 | 流式细胞术 | 80 | [ |
| 全菌 | Cell-SELEX | CCCCCGTTGCTTTCGCTTTTCCTTTCGCTTTTGTTCGTTTCGTCCCTGCTTCCTTTCTTG | 14.55 | 地高辛标记适配体 | 60 | [ |
| 丁酰基-高丝氨酸 | Structure-switching SELEX | CCATCCACACTCCGCAAGTGGGGAGGGGAGAGACGACGATCCTGTGGGTTTTCTGCAGTGAGTCGTGTTTTCGACTTATTGCGTCGGCTGCCTCTACAT | 28.47 | 流式细胞术 | 99 | [ |

Fig. 2 Aptamer sensor for P. aeruginosa detection A: Colorimetric biosensor for P. aeruginosa detection. B: Gold nanoparticle-based electrochemical biosensor for P. aeruginosa detection. C: Sandwich electrochemical biosensor for P. aeruginosa detection. D: Fluorescent biosensor for P. aeruginosa detection
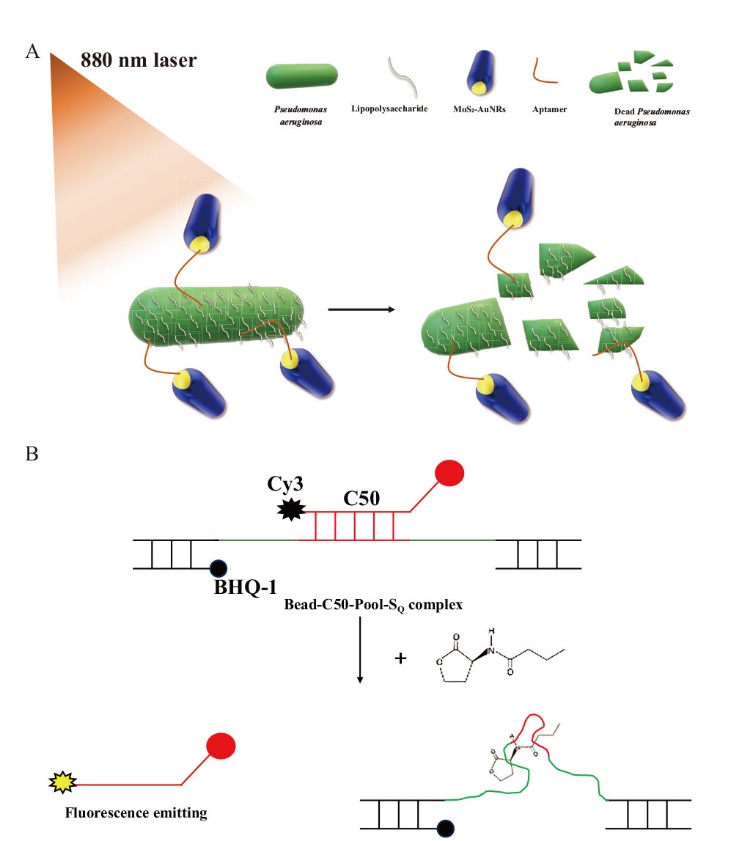
Fig. 3 Aptamer-based control of P. aeruginosa A: Aptamer-conjugated gold nanorods that inhibit P. aeruginosa. B: Aptamers that inhibit biofilm formation of P. aeruginosa
| [1] |
Panayidou S, Georgiades K, Christofi T, et al. Pseudomonas aeruginosa core metabolism exerts a widespread growth-independent control on virulence[J]. Sci Rep, 2020, 10(1): 9505.
doi: 10.1038/s41598-020-66194-4 pmid: 32528034 |
| [2] |
Hu PW, Chen JY, Chen YH, et al. Molecular epidemiology, resistance, and virulence properties of Pseudomonas aeruginosa cross-colonization clonal isolates in the non-outbreak setting[J]. Infect Genet Evol, 2017, 55: 288-296.
doi: 10.1016/j.meegid.2017.09.010 URL |
| [3] |
Ugwuanyi FC, Ajayi A, Ojo DA, et al. Evaluation of efflux pump activity and biofilm formation in multidrug resistant clinical isolates of Pseudomonas aeruginosa isolated from a Federal Medical Center in Nigeria[J]. Ann Clin Microbiol Antimicrob, 2021, 20(1): 11.
doi: 10.1186/s12941-021-00417-y |
| [4] |
Patel BM, Paratz JD, Mallet A, et al. Characteristics of bloodstream infections in burn patients: an 11-year retrospective study[J]. Burns, 2012, 38(5): 685-690.
doi: 10.1016/j.burns.2011.12.018 pmid: 22348799 |
| [5] |
Thi MTT, Wibowo D, Rehm BHA. Pseudomonas aeruginosa biofilms[J]. Int J Mol Sci, 2020, 21(22): 8671.
doi: 10.3390/ijms21228671 URL |
| [6] |
Jurado-Martín I, Sainz-Mejías M, McClean S. Pseudomonas aeruginosa: an audacious pathogen with an adaptable arsenal of virulence factors[J]. Int J Mol Sci, 2021, 22(6): 3128.
doi: 10.3390/ijms22063128 URL |
| [7] |
Sainz-Mejías M, Jurado-Martín I, McClean S. Understanding Pseudomonas aeruginosa-host interactions: the ongoing quest for an efficacious vaccine[J]. Cells, 2020, 9(12): 2617.
doi: 10.3390/cells9122617 URL |
| [8] |
Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands[J]. Nature, 1990, 346(6287): 818-822.
doi: 10.1038/346818a0 |
| [9] |
Qian SW, Chang DR, He SS, et al. Aptamers from random sequence space: Accomplishments, gaps and future considerations[J]. Anal Chim Acta, 2022, 1196: 339511.
doi: 10.1016/j.aca.2022.339511 URL |
| [10] |
Xu WT, He WC, Du ZH, et al. Functional nucleic acid nanomaterials: development, properties, and applications[J]. Angew Chem Int Ed Engl, 2021, 60(13): 6890-6918.
doi: 10.1002/anie.v60.13 URL |
| [11] |
李天顺, 李宸葳, 王佳, 等. 功能核酸筛选过程中次级文库的有效制备[J]. 生物技术通报, 2023, 39(3): 116-122.
doi: 10.13560/j.cnki.biotech.bull.1985.2022-0743 |
| Li TS, Li CW, Wang J, et al. Efficient generation of secondary libraries during functional nucleic acids screening[J]. Biotechnol Bull, 2023, 39(3): 116-122. | |
| [12] |
Qi S, Duan N, Khan IM, et al. Strategies to manipulate the performance of aptamers in SELEX, post-SELEX and microenvironment[J]. Biotechnol Adv, 2022, 55: 107902.
doi: 10.1016/j.biotechadv.2021.107902 URL |
| [13] |
Darmostuk M, Rimpelova S, Gbelcova H, et al. Current approaches in SELEX: an update to aptamer selection technology[J]. Biotechnol Adv, 2015, 33(6 Pt 2): 1141-1161.
doi: 10.1016/j.biotechadv.2015.02.008 pmid: 25708387 |
| [14] |
Wang KY, Zeng YL, Yang XY, et al. Utility of aptamer-fluorescence in situ hybridization for rapid detection of Pseudomonas aeruginosa[J]. Eur J Clin Microbiol Infect Dis, 2011, 30(2): 273-278.
doi: 10.1007/s10096-010-1074-0 URL |
| [15] |
Soundy J, Day D. Selection of DNA aptamers specific for live Pseudomonas aeruginosa[J]. PLoS One, 2017, 12(9): e0185385.
doi: 10.1371/journal.pone.0185385 URL |
| [16] | Hong KL, Yancey K, Battistella L, et al. Selection of single-stranded DNA molecular recognition elements against exotoxin A using a novel decoy-SELEX method and sensitive detection of exotoxin A in human serum[J]. Biomed Res Int, 2015, 2015: 417641. |
| [17] |
Ye H, Duan N, Gu HJ, et al. Fluorometric determination of lipopolysaccharides via changes of the graphene oxide-enhanced fluorescence polarization caused by truncated aptamers[J]. Mikrochim Acta, 2019, 186(3): 173.
doi: 10.1007/s00604-019-3261-8 pmid: 30771102 |
| [18] |
Davydova AS, Vorobyeva MA, Kabilov MR, et al. In vitro selection of cell-internalizing 2'-modified RNA aptamers against Pseudomonas aeruginosa[J]. Russ J Bioorg Chem, 2017, 43(1): 58-63.
doi: 10.1134/S1068162016060030 URL |
| [19] |
Wang HY, Chi Z, Cong Y, et al. Development of a fluorescence assay for highly sensitive detection of Pseudomonas aeruginosa based on an aptamer-carbon dots/graphene oxide system[J]. RSC Adv, 2018, 8(57): 32454-32460.
doi: 10.1039/C8RA04819C URL |
| [20] | 曾燕丽, 兰小鹏, 江丽, 等. 灭活铜绿假单胞菌适体的筛选[J]. 中国生物化学与分子生物学报, 2009, 25(1): 90-97. |
| Zeng YL, Lan XP, Jiang L, et al. Selection of aptamers specifically binding to inactivate Pseudomonas aeruginosa[J]. Chin J Biochem Mol Biol, 2009, 25(1): 90-97. | |
| [21] |
Zhao M, Li WB, Liu KC, et al. C4-HSL aptamers for blocking qurom sensing and inhibiting biofilm formation in Pseudomonas aeruginosa and its structure prediction and analysis[J]. PLoS One, 2019, 14(2): e0212041.
doi: 10.1371/journal.pone.0212041 URL |
| [22] |
Tang YJ, Ali Z, Zou J, et al. Detection methods for Pseudomonas aeruginosa: history and future perspective[J]. RSC Adv, 2017, 7(82): 51789-51800.
doi: 10.1039/C7RA09064A URL |
| [23] |
Deschaght P, Van Daele S, De Baets F, et al. PCR and the detection of Pseudomonas aeruginosa in respiratory samples of CF patients. A literature review[J]. J Cyst Fibros, 2011, 10(5): 293-297.
doi: 10.1016/j.jcf.2011.05.004 pmid: 21684819 |
| [24] |
Schmitz FRW, Cesca K, Valério A, et al. Colorimetric detection of Pseudomonas aeruginosa by aptamer-functionalized gold nanoparticles[J]. Appl Microbiol Biotechnol, 2023, 107(1): 71-80.
doi: 10.1007/s00253-022-12283-5 |
| [25] |
Das R, Dhiman A, Kapil A, et al. Aptamer-mediated colorimetric and electrochemical detection of Pseudomonas aeruginosa utilizing peroxidase-mimic activity of gold NanoZyme[J]. Anal Bioanal Chem, 2019, 411(6): 1229-1238.
doi: 10.1007/s00216-018-1555-z |
| [26] |
Abedi R, Bakhsh Raoof J, Mohseni M, et al. Sandwich-type electrochemical aptasensor for highly sensitive and selective detection of Pseudomonas aeruginosa bacteria using a dual signal amplification strategy[J]. Bioelectrochemistry, 2023, 150:108332.
doi: 10.1016/j.bioelechem.2022.108332 URL |
| [27] |
Roushani M, Sarabaegi M, Pourahmad F. Impedimetric aptasensor for Pseudomonas aeruginosa by using a glassy carbon electrode modified with silver nanoparticles[J]. Mikrochim Acta, 2019, 186(11): 725.
doi: 10.1007/s00604-019-3858-y pmid: 31655899 |
| [28] |
Xie YX, Xie GM, Yuan JS, et al. A novel fluorescence biosensor based on double-stranded DNA branch migration-induced HCR and DNAzyme feedback circuit for sensitive detection of Pseudomonas aeruginosa(clean version)[J]. Anal Chim Acta, 2022, 1232: 340449.
doi: 10.1016/j.aca.2022.340449 URL |
| [29] |
Hu JY, Fu KY, Bohn PW. Whole-cell Pseudomonas aeruginosa localized surface plasmon resonance aptasensor[J]. Anal Chem, 2018, 90(3): 2326-2332.
doi: 10.1021/acs.analchem.7b04800 URL |
| [30] |
Zhong ZT, Gao XM, Gao R, et al. Selective capture and sensitive fluorometric determination of Pseudomonas aeruginosa by using aptamer modified magnetic nanoparticles[J]. Mikrochim Acta, 2018, 185(8): 377.
doi: 10.1007/s00604-018-2914-3 |
| [31] |
Mayr FB, Yende S, Angus DC. Epidemiology of severe sepsis[J]. Virulence, 2014, 5(1): 4-11.
doi: 10.4161/viru.27372 pmid: 24335434 |
| [32] |
Li J, Zhang YZ, Zhu LJ, et al. Smart nucleic acid hydrogels with high stimuli-responsiveness in biomedical fields[J]. Int J Mol Sci, 2022, 23(3): 1068.
doi: 10.3390/ijms23031068 URL |
| [33] |
Yang M, Chen X, Zhu LJ, et al. Aptamer-functionalized DNA-silver nanocluster nanofilm for visual detection and elimination of bacteria[J]. ACS Appl Mater Interfaces, 2021, 13(32): 38647-38655.
doi: 10.1021/acsami.1c05751 URL |
| [34] |
Rolband L, Yourston L, Chandler M, et al. DNA-templated fluorescent silver nanoclusters inhibit bacterial growth while being non-toxic to mammalian cells[J]. Molecules, 2021, 26(13): 4045.
doi: 10.3390/molecules26134045 URL |
| [35] | Lv J, Li B, Luo TT, et al. Selective photothermal therapy based on lipopolysaccharide aptamer functionalized MoS2 nanosheet-coated gold nanorods for multidrug-resistant Pseudomonas aeruginosa infection[J]. Adv Healthc Mater, 2023, 12(15): e2202794. |
| [36] |
Kraemer M, Bellion M, Kissmann AK, et al. Aptamers as novel binding molecules on an antimicrobial peptide-armored composite hydrogel wound dressing for specific removal and efficient eradication of Pseudomonas aeruginosa[J]. Int J Mol Sci, 2023, 24(5):4800.
doi: 10.3390/ijms24054800 URL |
| [37] |
Soundy J, Day D. Delivery of antibacterial silver nanoclusters to Pseudomonas aeruginosa using species-specific DNA aptamers[J]. J Med Microbiol, 2020, 69(4): 640-652.
doi: 10.1099/jmm.0.001174 URL |
| [1] | FENG Lu-yao, ZHAO Jiang-yuan, SHI Zhu-feng, MO Yan-fang, YANG Tong-yu, SHEN Yun-xin, HE Fei-fei, LI Ming-gang, YANG Pei-wen. Isolation and Identification of Bacteria in Forest Rhizosphere Soil and Their Biological Activity Screening [J]. Biotechnology Bulletin, 2024, 40(1): 294-307. |
| [2] | HUANG Xiao-long, SUN Gui-lian, MA Dan-dan, YAN Hui-qing. Construction of Yeast One-hybrid Library and Screening of Factors Regulating LAZY1 Expression in Rice [J]. Biotechnology Bulletin, 2023, 39(9): 126-135. |
| [3] | WEN Xiao-lei, LI Jian-yuan, LI Na, ZHANG Na, YANG Wen-xiang. Construction and Utilization of Yeast Two-hybrid cDNA Library of Wheat Interacted by Puccinia triticina [J]. Biotechnology Bulletin, 2023, 39(9): 136-146. |
| [4] | WU Qiao-yin, SHI You-zhi, LI Lin-lin, PENG Zheng, TAN Zai-yu, LIU Li-ping, ZHANG Juan, PAN Yong. In Situ Screening of Carotenoid Degrading Strains and the Application in Improving Quality and Aroma of Cigar [J]. Biotechnology Bulletin, 2023, 39(9): 192-201. |
| [5] | JIANG Hai-rong, CUI Ruo-qi, WANG Yue BAI, Miao ZHANG, Ming-lu , REN Lian-hai. Isolation, Identification and Degradation Characteristics of Functional Bacteria for NH3 and H2S Degradation [J]. Biotechnology Bulletin, 2023, 39(9): 246-254. |
| [6] | XUE Ning, WANG Jin, LI Shi-xin, LIU Ye, CHENG Hai-jiao, ZHANG Yue, MAO Yu-feng, WANG Meng. Construction of L-phenylalanine High-producing Corynebacterium glutamicum Engineered Strains via Multi-gene Simultaneous Regulation Combined with High-throughput Screening [J]. Biotechnology Bulletin, 2023, 39(9): 268-280. |
| [7] | XU Fa-di, XU Kang, SUN Dong-ming, LI Meng-lei, ZHAO Jian-zhi, BAO Xiao-ming. Research Progress in Second-generation Fuel Ethanol Technology Based on Poplar(Populus sp.) [J]. Biotechnology Bulletin, 2023, 39(9): 27-39. |
| [8] | MA Jun-xiu, WU Hao-qiong, JIANG Wei, YAN Geng-xuan, HU Ji-hua, ZHANG Shu-mei. Screening and Identification of Broad-spectrum Antagonistic Bacterial Strains Against Vegetable Soft Rot Pathogen and Its Control Effects [J]. Biotechnology Bulletin, 2023, 39(7): 228-240. |
| [9] | XIE Dong, WANG Liu-wei, LI Ning-jian, LI Ze-lin, XU Zi-hang, ZHANG Qing-hua. Exploration, Identification and Phosphorus-solubilizing Condition Optimization of a Multifunctional Strain [J]. Biotechnology Bulletin, 2023, 39(7): 241-253. |
| [10] | YUAN Ye, ZHOU Jia, QU Jian-hang, ZHANG Bo-yuan, LUO Yu, LI Hai-feng. Screening of an Efficient Denitrifying Phosphorus-accumulating Bacterium and Its Denitrification and Phosphorus Removal [J]. Biotechnology Bulletin, 2023, 39(7): 266-276. |
| [11] | YOU Zi-juan, CHEN Han-lin, DENG Fu-cai. Research Progress in the Extraction and Functional Activities of Bioactive Peptides from Fish Skin [J]. Biotechnology Bulletin, 2023, 39(7): 91-104. |
| [12] | LI Dian-dian, SU Yuan, LI Jie, XU Wen-tao, ZHU Long-jiao. Progress in Selection and Application of Antibacterial Aptamers [J]. Biotechnology Bulletin, 2023, 39(6): 126-132. |
| [13] | DONG Cong, GAO Qing-hua, WANG Yue, LUO Tong-yang, WANG Qing-qing. Increasing the Expression of FAD-dependent Glucose Dehydrogenase by Recombinant Pichia pastoris Using a Combined Strategy [J]. Biotechnology Bulletin, 2023, 39(6): 316-324. |
| [14] | LI Zhi-qi, YUAN Yue, MIAO Rong-qing, PANG Qiu-ying, ZHANG Ai-qin. Melatonin Contents in Eutrema salsugineum and Arabidopsis thaliana Under Salt Stress, and Expression Pattern Analysis of Synthesis Related Genes [J]. Biotechnology Bulletin, 2023, 39(5): 142-151. |
| [15] | ZHANG Xue-ping, LU Yu-qing, ZHANG Yue-qian, LI Xiao-juan. Advances in Plant Extracellular Vesicles and Analysis Techniques [J]. Biotechnology Bulletin, 2023, 39(5): 32-43. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||