








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (5): 215-224.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1139
Previous Articles Next Articles
LI Jing-yan1( ), ZHOU Jia-jing2, YUAN Yuan3, SU Xiao-yi1, QIAO Wen-hui2, XUE Yan-lei3, LI Guo-jing1, WANG Rui-gang1(
), ZHOU Jia-jing2, YUAN Yuan3, SU Xiao-yi1, QIAO Wen-hui2, XUE Yan-lei3, LI Guo-jing1, WANG Rui-gang1( )
)
Received:2023-12-03
Online:2024-05-26
Published:2024-06-13
Contact:
WANG Rui-gang
E-mail:3265729074@qq.com;wangruigang@imau.edu.cn
LI Jing-yan, ZHOU Jia-jing, YUAN Yuan, SU Xiao-yi, QIAO Wen-hui, XUE Yan-lei, LI Guo-jing, WANG Rui-gang. Response of Arabidopsis AtiPGAM2 Gene to Abiotic Stress[J]. Biotechnology Bulletin, 2024, 40(5): 215-224.
| 引物名称Primer name | 序列Sequence(5'-3') | 引物用途Primer usage |
|---|---|---|
| AtiPGAM2-HA-F | TACTTCCAATCCAATGCCATGGGTAGCTCCGGCG | 基因克隆 |
| AtiPGAM2-HA-R | TTATCCACTTCCAATGCTACTTCTCGACGACTTCGATCAG | Gene clone |
| AtiPGAM2-qRT-F | TGCGTGTAAACCTGCCAAAT | 荧光定量PCR |
| AtiPGAM2-qRT-R | GTATATCCCTCCGACCTGTTCTAT | RT-qPCR |
| AtEF1α-F | AGAAGGGTGCCAAATGATGAG | 荧光定量PCR |
| AtEF1α-R | GGAGGGAGAGAGAAAGTCACAGA | RT-qPCR |
| salk_119825C-LP | TACCGAACCAGATCAATTTGC | 突变体鉴定 |
| salk_119825C-RP | TCCTTGATGCCATAGAACAGG | Mutant identification |
Table 1 Primers used in this study
| 引物名称Primer name | 序列Sequence(5'-3') | 引物用途Primer usage |
|---|---|---|
| AtiPGAM2-HA-F | TACTTCCAATCCAATGCCATGGGTAGCTCCGGCG | 基因克隆 |
| AtiPGAM2-HA-R | TTATCCACTTCCAATGCTACTTCTCGACGACTTCGATCAG | Gene clone |
| AtiPGAM2-qRT-F | TGCGTGTAAACCTGCCAAAT | 荧光定量PCR |
| AtiPGAM2-qRT-R | GTATATCCCTCCGACCTGTTCTAT | RT-qPCR |
| AtEF1α-F | AGAAGGGTGCCAAATGATGAG | 荧光定量PCR |
| AtEF1α-R | GGAGGGAGAGAGAAAGTCACAGA | RT-qPCR |
| salk_119825C-LP | TACCGAACCAGATCAATTTGC | 突变体鉴定 |
| salk_119825C-RP | TCCTTGATGCCATAGAACAGG | Mutant identification |
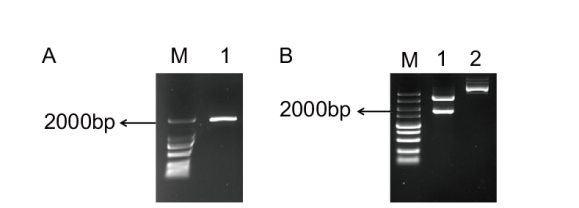
Fig. 1 Gel electrophoresis of the PCR products of AtiP-GAM2 ORF cloned(A)and identification of 35S:: AtiPGAM2-HA recombinant vector(B) A: M: DL2000 DNA marker; 1: PCR products of the AtiPGAM2 ORF; B: M: DL2000 DNA marker; 1: recombinant vector digested with Sal I; 2: vector control
| 顺式作用元件名称 Name of cis-acting element | 顺式作用元件功能 Function of cis-acting element | 序列 Sequence(5'-3') | 顺式作用元件数量 Number of cis-acting elements |
|---|---|---|---|
| TGACG-motif | MeJA反应相关 | TGACG | 4 |
| CGTCA-motif | MeJA反应相关 | CGTCA | 4 |
| G-box | 光反应相关 | CACGTC | 3 |
| G-box | 光反应相关 | CACGTT | 1 |
| ABRE | 脱落酸反应相关 | ACGTG | 4 |
| TCA-element | 水杨酸反应相关 | TCAGAAGAGG | 1 |
| TCA-element | 水杨酸反应相关 | CCATCTTTTT | 1 |
| TCCC-motif | 光反应相关 | TCTCCCT | 1 |
| P-box | 赤霉素相关 | CCTTTTG | 2 |
| I-box | 光反应相关 | CCTTATCCT | 1 |
| GARE-motif | 赤霉素响应元件 | TCTGTTG | 1 |
| GT1-motif | 光响应元件 | GGTTAA | 1 |
| GATA-motif | 光反应相关 | GATAGGA | 1 |
| GATA-motif | 光反应相关 | AAGGATAAGG | 1 |
| LTR | 低温响应元件 | CCGAAA | 1 |
| TCT-motif | 光响应元件 | TCTTAC | 2 |
Table 2 Cis-acting elements in AtiPGAM2 promoter
| 顺式作用元件名称 Name of cis-acting element | 顺式作用元件功能 Function of cis-acting element | 序列 Sequence(5'-3') | 顺式作用元件数量 Number of cis-acting elements |
|---|---|---|---|
| TGACG-motif | MeJA反应相关 | TGACG | 4 |
| CGTCA-motif | MeJA反应相关 | CGTCA | 4 |
| G-box | 光反应相关 | CACGTC | 3 |
| G-box | 光反应相关 | CACGTT | 1 |
| ABRE | 脱落酸反应相关 | ACGTG | 4 |
| TCA-element | 水杨酸反应相关 | TCAGAAGAGG | 1 |
| TCA-element | 水杨酸反应相关 | CCATCTTTTT | 1 |
| TCCC-motif | 光反应相关 | TCTCCCT | 1 |
| P-box | 赤霉素相关 | CCTTTTG | 2 |
| I-box | 光反应相关 | CCTTATCCT | 1 |
| GARE-motif | 赤霉素响应元件 | TCTGTTG | 1 |
| GT1-motif | 光响应元件 | GGTTAA | 1 |
| GATA-motif | 光反应相关 | GATAGGA | 1 |
| GATA-motif | 光反应相关 | AAGGATAAGG | 1 |
| LTR | 低温响应元件 | CCGAAA | 1 |
| TCT-motif | 光响应元件 | TCTTAC | 2 |
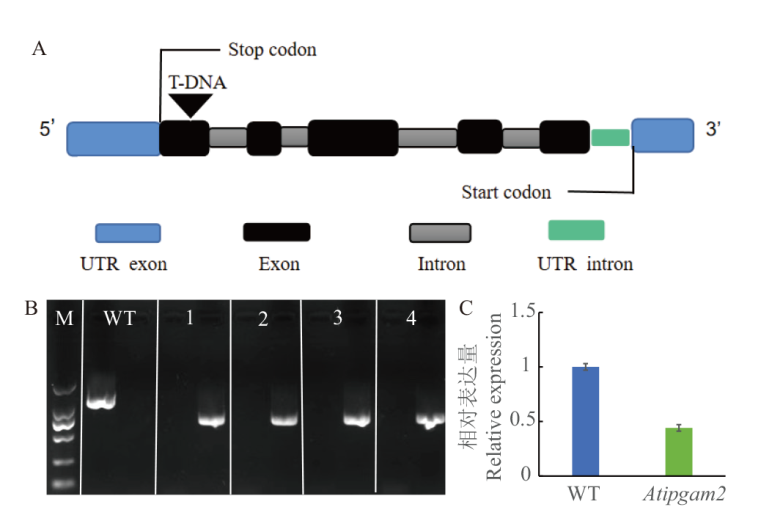
Fig. 4 T-DNA insertion site in mutant Atipgam2 and identification of its expression A: T-DNA insertion site of salk_119825C mutant ; B: PCR identification of salk_119825C mutant(M: DL 2000 marker; 1-4: the number of Atipgam2 mutant; 1-4 left swimming lane: PCR products of LP+RP; 1-4 right swimming lane: PCR products of BP+ RP); C: detection of AtiPGAM2 gene expression levels by RT qPCR
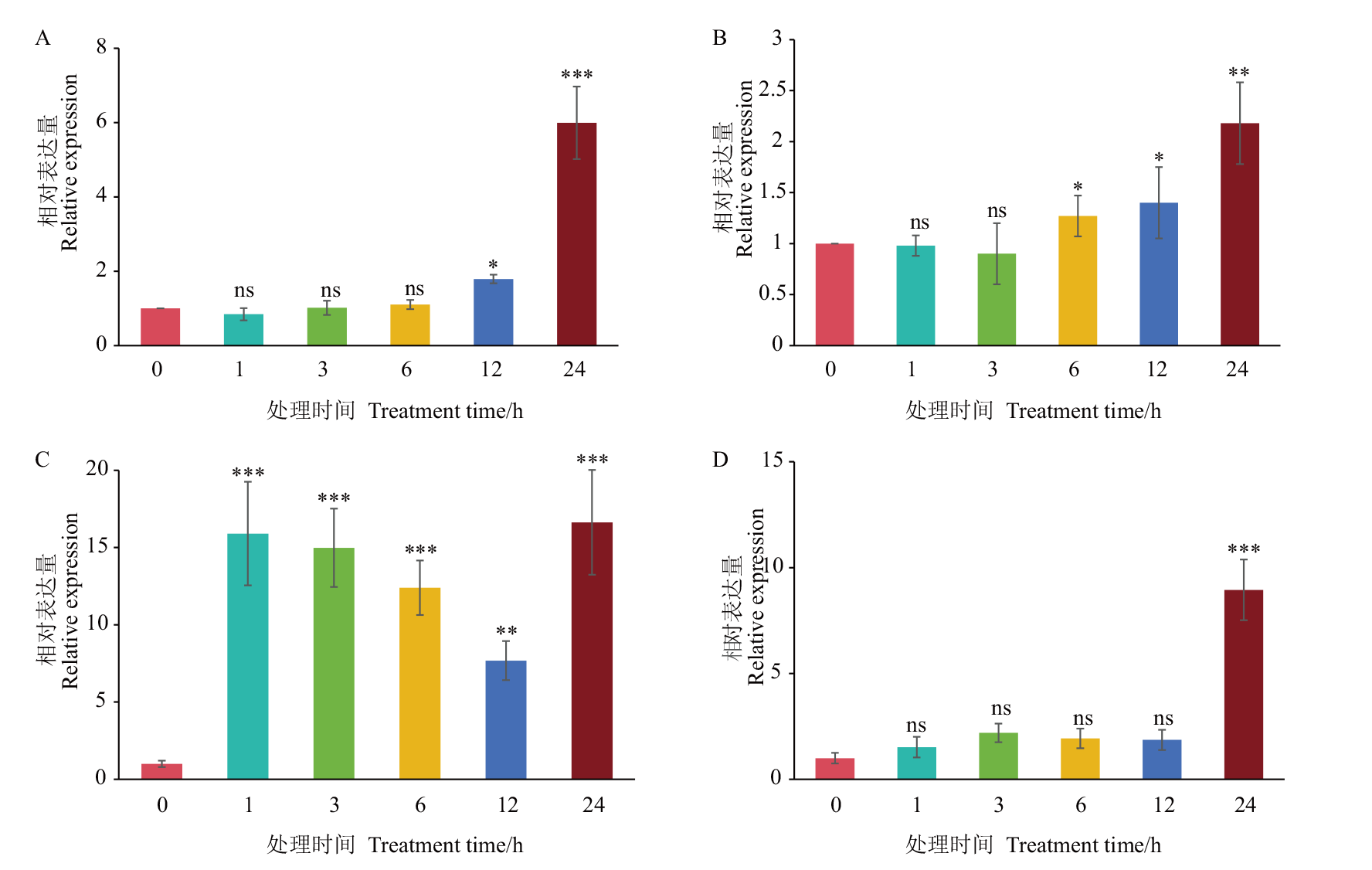
Fig. 5 Transcriptional changes of AtiPGAM2 under salt stress and ABA A: Transcription level change of RD29A under salt stress. B: Transcription level change of AtiPGAM2 under salt stress. C: Transcription level change of ABI1 under ABA. D: Transcription level change of AtiPGAM2 under ABA. *P<0.05,** P<0.01, *** P<0.001, ns refers to insignificant. The same below

Fig. 6 Effects of NaCl on the germination rate and green seedling rate of wild-type, AtiPGAM2 overexpressed homozygotes and Atipgam2 mutant A, C: Phenotypes(A)and germination rate(C)of Arabidopsis seeds of three genotypes under control conditions after 7 d of germination. B, D: Phenotypes(B)and germination rate(D)of Arabidopsis seeds of three genotypes under 200 mmol/L NaCl treatment after 7 d of germination. E: Phenotypes of green seedling rates of three Arabidopsis genotypes under control after 10 d of germination. F: Phenotypes of green seedling rates of three Arabidopsis genotypes under 150 mmol/L NaCl treatment after 10 d of germination. G: Statistical results of the green seedling rates of Arabidopsis of three genotypes under control and 150 mmol/L NaCl treatment after 10 d of germination

Fig. 7 Effects of mannitol on the germination rate of wild-type, AtiPGAM2 overexpressed homozygotes and Atipgam2 mutant A, C: Phenotypes(A)and germination rate(C)of Arabidopsis seeds of three genotypes under control conditions after 7 d of germination. B, D: Phenotypes(B)and germination rate(D)of Arabidopsis seeds of three genotypes under 400 mmol/L mannitol treatment after 7 d of germination

Fig. 8 Effects of ABA on the germination rate of wild-type, AtiPGAM2 overexpressed homozygotes and Atipgam2 mutant A, D: Phenotypes(A)and germination rate(D)of Arabidopsis seeds of three genotypes under control conditions after 7 d of germination. B, E: Phenotypes(B)and germination rate(E)of Arabidopsis seeds of three genotypes under 1 μmol /L ABA treatment after 7 d of germination. C, F: Phenotypes(C)and germination rate(F)of Arabidopsis seeds of three genotypes under 3 μmol/L ABA treatment after 7 d of germination
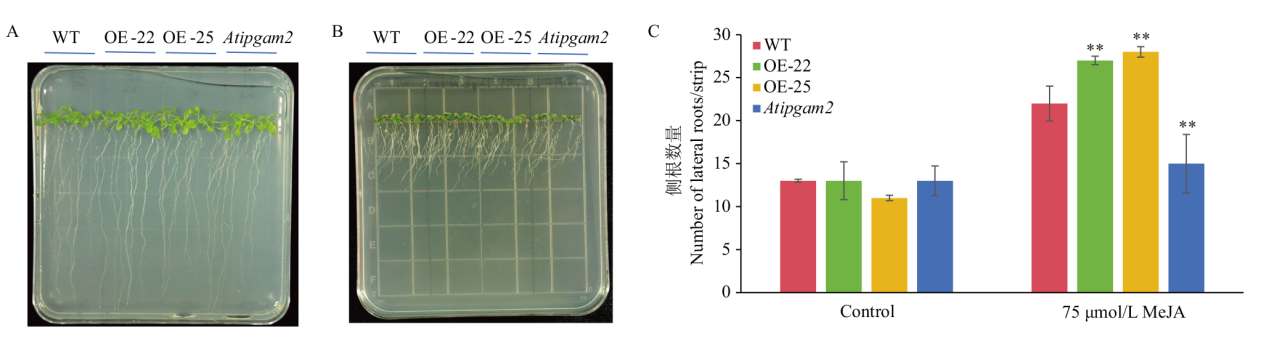
Fig. 9 Effects of MeJA on the number of lateral roots in wild-type, AtiPGAM2 overexpressed homozygotes, and Atipgam2 mutants A: Phenotypes of lateral root number of three Arabidopsis genotypes under control. B: Phenotypes of lateral root number of three Arabidopsis genotypes under 75 μmol/L MeJA treatment. C: Statistical results of lateral root average number of Arabidopsis of three genotypes under control and 75 μmol/L MeJA treatment
| [1] | Okuda J, Niizuma S, Shioi T, et al. Persistent overexpression of phosphoglycerate mutase, a glycolytic enzyme, modifies energy metabolism and reduces stress resistance of heart in mice[J]. PLoS One, 2013, 8(8): e72173. |
| [2] |
Fothergill-Gilmore LA, Watson HC. The phosphoglycerate mutases[J]. Adv Enzymol Relat Areas Mol Biol, 1989, 62: 227-313.
pmid: 2543188 |
| [3] | Jedrzejas MJ. Structure, function, and evolution of phosphoglycerate mutases: comparison with fructose-2, 6-bisphosphatase, acid phosphatase, and alkaline phosphatase[J]. Prog Biophys Mol Biol, 2000, 73(2/3/4): 263-287. |
| [4] | Poonperm B, Guerra DG, McNae IW, et al. Expression, purification, crystallization and preliminary crystallographic analysis of Leishmania mexicana phosphoglycerate mutase[J]. Acta Crystallogr D Biol Crystallogr, 2003, 59(Pt 7): 1313-1316. |
| [5] |
Wang YL, Wei ZY, Bian Q, et al. Crystal structure of human bisphosphoglycerate mutase[J]. J Biol Chem, 2004, 279(37): 39132-39138.
doi: 10.1074/jbc.M405982200 pmid: 15258155 |
| [6] |
谢鑫, 刘娜, 魏凤菊. 植物iPGAM的研究进展[J]. 生物技术通报, 2015, 31(9): 1-7.
doi: 10.13560/j.cnki.biotech.bull.1985.2015.09.001 |
|
Xie X, Liu N, Wei FJ. Research progress on cofactor-independent phosphoglycerate mutase in plants[J]. Biotechnol Bull, 2015, 31(9): 1-7.
doi: 10.13560/j.cnki.biotech.bull.1985.2015.09.001 |
|
| [7] | Graña X, Pérez de la Ossa P, Broceño C, et al. 2, 3-Bisphosphoglycerate-independent phosphoglycerate mutase is conserved among different phylogenic Kingdoms[J]. Comp Biochem Physiol B Biochem Mol Biol, 1995, 112(2): 287-293. |
| [8] |
Bond CS, Clements PR, Ashby SJ, et al. Structure of a human lysosomal sulfatase[J]. Structure, 1997, 5(2): 277-289.
doi: 10.1016/s0969-2126(97)00185-8 pmid: 9032078 |
| [9] |
Andriotis VME, Kruger NJ, Pike MJ, et al. Plastidial glycolysis in developing Arabidopsis embryos[J]. New Phytol, 2010, 185(3): 649-662.
doi: 10.1111/j.1469-8137.2009.03113.x pmid: 20002588 |
| [10] | Graña X, Ureña J, Ludevid D, et al. Purification, characterization and immunological properties of 2, 3-bisphosphoglycerate-independent phosphoglycerate mutase from maize(Zea mays)seeds[J]. Eur J Biochem, 1989, 186(1/2): 149-153. |
| [11] |
Wang JL, Walling LL, Jauh GY, et al. Lily cofactor-independent phosphoglycerate mutase: purification, partial sequencing, and immunolocalization[J]. Planta, 1996, 200(3): 343-352.
pmid: 8931352 |
| [12] | Westram A, Lloyd JR, Roessner U, et al. Increases of 3-phosphoglyceric acid in potato plants through antisense reduction of cytoplasmic phosphoglycerate mutase impairs photosynthesis and growth, but does not increase starch contents[J]. Plant Cell Environ, 2002, 25(9): 1133-1143. |
| [13] | Lin D, Zhang L, Mei J, et al. Mutation of the rice TCM12 gene encoding 2, 3-bisphosphoglycerate-independent phosphoglycerate mutase affects chlorophyll synthesis, photosynthesis and chloroplast development at seedling stage at low temperatures[J]. Plant Biol, 2019, 21(4): 585-594. |
| [14] | 刘朋虎, 邓优锦, 江玉姬, 等. 草菇PGAM基因克隆、结构及其在同核、异核菌株中的表达量分析[J]. 福建农业学报, 2012, 27(3): 252-256. |
| Liu PH, Deng YJ, Jiang YJ, et al. Cloning and structural analysis of phosphoglycerate mutas gene and its expression levels in homokaryon and heterokaryon of Volvariella volvacea[J]. Fujian J Agric Sci, 2012, 27(3): 252-256. | |
| [15] | 李春明, 白卉, 于文喜. 低温驯化过程中大青杨叶片差异蛋白质分析[J]. 东北林业大学学报, 2011, 39(10): 45-49. |
| Li CM, Bai H, Yu WX. Differential protein in Populus ussuriensis leaves during natural low-temperature acclimation by proteomic analysis[J]. J Northeast For Univ, 2011, 39(10): 45-49. | |
| [16] |
Huang Y, Blakeley SD, McAleese SM, et al. Higher-plant cofactor-independent phosphoglyceromutase: purification, molecular characterization and expression[J]. Plant Mol Biol, 1993, 23(5): 1039-1053.
doi: 10.1007/BF00021818 pmid: 8260624 |
| [17] |
Hajduch M, Casteel JE, Hurrelmeyer KE, et al. Proteomic analysis of seed filling in Brassica napus. Developmental characterization of metabolic isozymes using high-resolution two-dimensional gel electrophoresis[J]. Plant Physiol, 2006, 141(1): 32-46.
pmid: 16543413 |
| [18] | 龙翔宇, 董绪浓, 等. 橡胶树PGAM基因的克隆及表达特性分析[J]. 热带作物学报, 2013, 34(10): 1895-1901. |
| Long XY, Dong XN, et al. Molecular cloning and expression analysis of HbPGAM from Hevea brasiliensis(para rubber tree)[J]. Chin J Trop Crops, 2013, 34(10): 1895-1901. | |
| [19] |
Stein M, Gabdoulline RR, Wade RC. Cross-species analysis of the glycolytic pathway by comparison of molecular interaction fields[J]. Mol Biosyst, 2010, 6(1): 152-164.
doi: 10.1039/b912398a pmid: 20024078 |
| [20] | Zhao ZX, Assmann SM. The glycolytic enzyme, phosphoglycerate mutase, has critical roles in stomatal movement, vegetative growth, and pollen production in Arabidopsis thaliana[J]. J Exp Bot, 2011, 62(14): 5179-5189. |
| [21] | Amme S, Matros A, Schlesier B, et al. Proteome analysis of cold stress response in Arabidopsis thaliana using DIGE-technology[J]. J Exp Bot, 2006, 57(7): 1537-1546. |
| [22] | 红格日其其格. 盐胁迫下拟南芥角果的蛋白质组和Khib修饰组学分析及AtATPase-β亚基的功能研究[D]. 呼和浩特: 内蒙古农业大学, 2021. |
| Hong G. Proteomic and Khib modification analysis of Arabidopsis thaliana siliques under salt stress and function study on ATATPase-β subunit[D]. Hohhot: Inner Mongolia Agricultural University, 2021. | |
| [23] | Zhang XJ, Yang GY, et al. Arabidopsis cysteine-rich receptor-like kinase 45 functions in the responses to abscisic acid and abiotic stresses[J]. Plant Physiol Biochem, 2013, 67: 189-198. |
| [24] | Karimi R, Gavili-Kilaneh K, Khadivi A. Methyl jasmonate promotes salinity adaptation responses in two grapevine(Vitis vinifera L.) cultivars differing in salt tolerance[J]. Food Chem, 2022, 375: 131667. |
| [25] | 刘丽婧. 烟草糖酵解途径iPGAM基因的鉴定和功能研究[D]. 重庆: 西南大学, 2020. |
| Liu LJ. Identification and functional study of NtiPGAM involved in glycolytic pathway in tobacco[D]. Chongqing: Southwest University, 2020. |
| [1] | DU Bing-shuai, ZOU Xin-hui, WANG Zi-hao, ZHANG Xin-yuan, CAO Yi-bo, ZHANG Ling-yun. Genome-wide Identification and Expression Analysis of the SWEET Gene Family in Camellia oleifera [J]. Biotechnology Bulletin, 2024, 40(5): 179-190. |
| [2] | GUO Hui-yan, DONG Xue, AN Meng-nan, XIA Zi-hao, WU Yuan-hua. Research Progress in the Functions of Key Enzymes of Ubiquitination Modification in Plant Stress Responses [J]. Biotechnology Bulletin, 2024, 40(4): 1-11. |
| [3] | LI Hui, WEN Yu-fang, WANG Yue, JI Chao, SHI Guo-you, LUO Ying, ZHOU Yong, LI Zhi-min, WU Xiao-yu, YANG You-xin, LIU Jian-ping. Expression Characteristics and Functions of CaPIF4 in Capsicum annuum Under Salt Stress [J]. Biotechnology Bulletin, 2024, 40(4): 148-158. |
| [4] | GUO Chun, SONG Gui-mei, YAN Yan, DI Peng, WANG Ying-ping. Genome Wide Identification and Expression Analysis of the bZIP Gene Family in Panax quinquefolius [J]. Biotechnology Bulletin, 2024, 40(4): 167-178. |
| [5] | GAO Yu-kun, ZHANG Jian-dong, YANG Pu-yuan, CHEN Dong-ming, WANG Zhi-bo, TIAN Yi-jin, Zakey Eldinn. E. A. Khlid, CUI Jiang-hui, CHANG Jin-hua. Responses of Sorghum Rhizosphere Soil Bacterial Communities to Salt Stress [J]. Biotechnology Bulletin, 2024, 40(4): 203-216. |
| [6] | GAO Zhi-wei, WEI Ming, YU Zu-long, WU Guo-qiang, WEI Jun-long. Identification of Salt-tolerant Plant Growth-promoting Bacterium W-1 and Its Effect on the Salt-tolerance of Sainfoin(Onobrychis viciaefolia) [J]. Biotechnology Bulletin, 2024, 40(4): 217-227. |
| [7] | JIANG Lin-qi, ZHAO Jia-ying, ZHENG Fei-xiong, YAO Xin-yi, LI Xiao-xian, YU Zhen-ming. Identification and Expression Analysis of 14-3-3 Gene Family in Dendrobium officinale [J]. Biotechnology Bulletin, 2024, 40(3): 229-241. |
| [8] | ZHOU Hong-dan, LUO Xiao-ping, TU Mi-xue, LI Zhong-guang. Phytomelatonin: An Emerging Signal Molecule Responding to Abiotic Stress [J]. Biotechnology Bulletin, 2024, 40(3): 41-51. |
| [9] | SHEN Tian-hong, QI Xiao-bo, ZHAO Rui-feng, MA Xin-rong. Research Progress in the Molecular Mechanisms of Microalgae Responding to Salt Stress [J]. Biotechnology Bulletin, 2024, 40(3): 89-99. |
| [10] | WU Cui-cui, XIAO Shui-ping. Genome-wide Identification of HD-Zip Gene Family in Gossypium hirsutum L. and Expression Analysis in Response to Abiotic Stress [J]. Biotechnology Bulletin, 2024, 40(2): 130-145. |
| [11] | XIN Qi, LI Ya-fan, YIN Zheng, ZHANG Xiao-dan, CHEN Ting, LIU Xiao-hua. Identification and Expression Analysis of the CBL-CIPK Gene Family in Sugarcane [J]. Biotechnology Bulletin, 2024, 40(2): 197-211. |
| [12] | LI Hao, WU Guo-qiang, WEI Ming, HAN Yue-xin. Genome-wide Identification of the BvBADH Gene Family in Sugar Beet(Beta vulgaris)and Their Expression Analysis Under High Salt Stress [J]. Biotechnology Bulletin, 2024, 40(2): 233-244. |
| [13] | XU Yang, ZHANG Rui-ying, DAI Liang-xiang, ZHANG Guan-chu, DING Hong, ZHANG Zhi-meng. Regulation of Nitrogen Application on Peanut Seed Germination and Spermosphere Bacterial Community Structure Under Salt Stress [J]. Biotechnology Bulletin, 2024, 40(2): 253-265. |
| [14] | LI Ya-nan, ZHANG Hao-jie, LIANG Meng-jing, LUO Tao, LI Wang-ning, ZHANG Chun-hui, JI Chun-li, LI Run-zhi, XUE Jin-ai, CUI Hong-li. Identification and Expression Analysis of Calcium-dependent Protein Kinase(CDPK)Family in Haematococcus pluvialis [J]. Biotechnology Bulletin, 2024, 40(2): 300-312. |
| [15] | ZOU Xiu-wei, YUE Jia-ni, LI Zhi-yu, DAI Liang-ying, LI Wei. Functional Analysis of Rice Heat Shock Transcription Factor HsfA2b Regulating the Resistance to Abiotic Stresses [J]. Biotechnology Bulletin, 2024, 40(2): 90-98. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||