








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (5): 248-260.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1174
Previous Articles Next Articles
SUN Ya-nan( ), WANG Chun-xue, WANG Xin, DU Bing-hai, LIU Kai(
), WANG Chun-xue, WANG Xin, DU Bing-hai, LIU Kai( ), WANG Cheng-qiang(
), WANG Cheng-qiang( )
)
Received:2023-12-13
Online:2024-05-26
Published:2024-03-21
Contact:
LIU Kai, WANG Cheng-qiang
E-mail:sun15383646425@163.com;liukai_1982@163.com;wangcq@sdau.edu.cn
SUN Ya-nan, WANG Chun-xue, WANG Xin, DU Bing-hai, LIU Kai, WANG Cheng-qiang. Biocontrol Characteristics of Bacillus atrophaeus CNY01 and Its Salt-resistant and Growth-promoting Effect on Maize Seedling[J]. Biotechnology Bulletin, 2024, 40(5): 248-260.

Fig. 3 Determination of salt and acid-alkali tolerance of strain CNY01 A: Growth of CNY01 on solid medium with different salt concentrations; B: salt tolerance test of CNY01 in liquid medium; C: determination of acid and alkali resistance of CNY01 in liquid medium
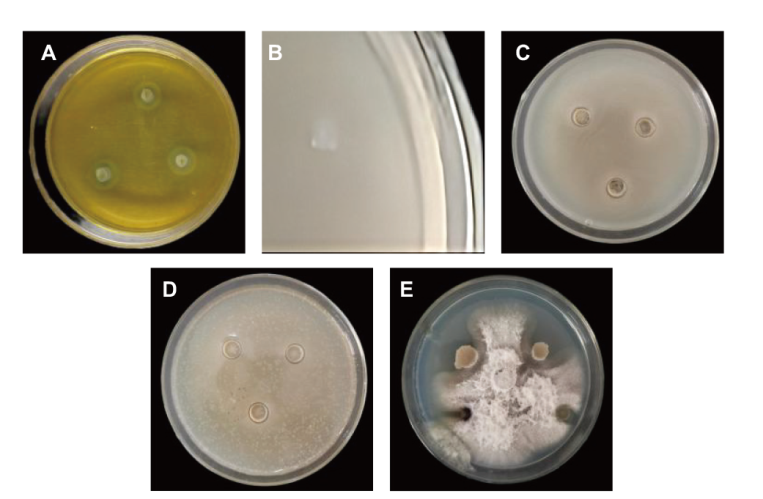
Fig. 4 Determination of antagonistic and growth-promoting ability of strain CNY01 A: CNY01 cultured in casein medium; B: CNY01 cultured in silicate medium; C: CNY01 cultured in antagonism with Bacillus subtilis; D: CNY01 cultured in antagonism with Ralstonia solanacearum; E: CNY01 cultured in antagonism with Fusarium moniliforme
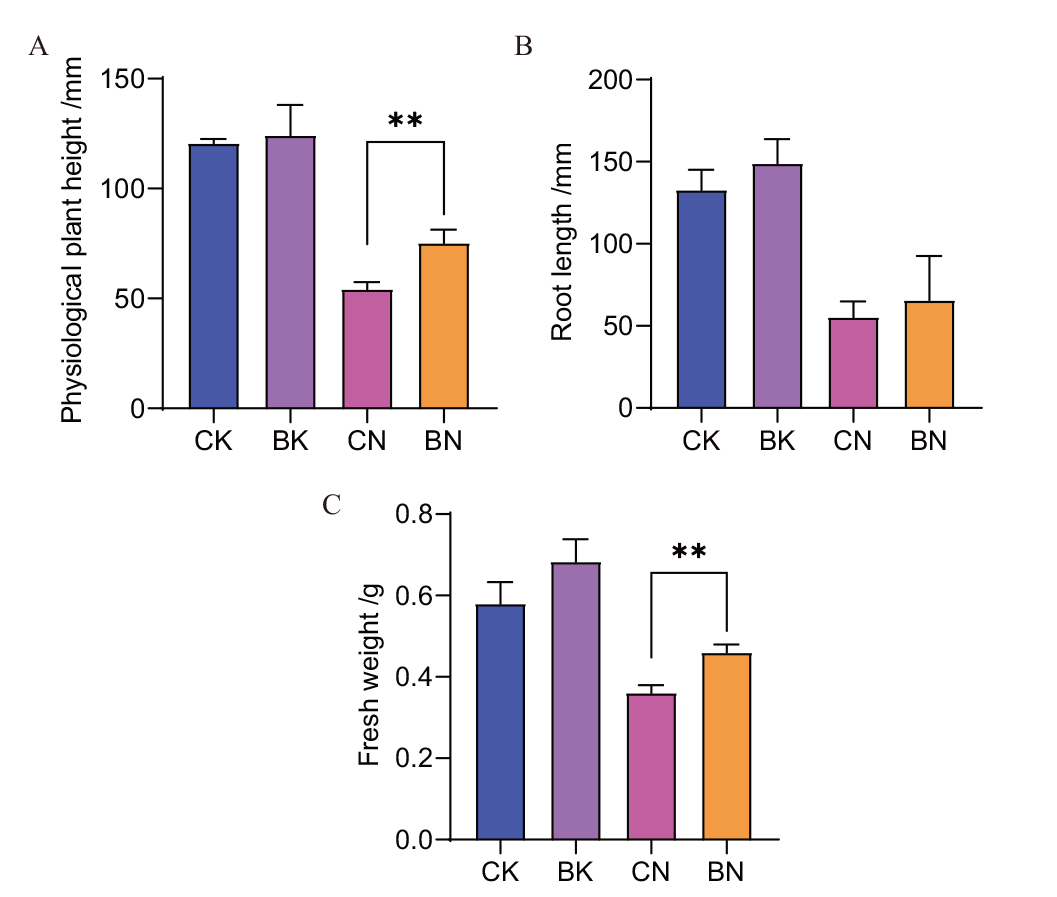
Fig. 5 Growth-promoting effect of strain CNY01 on maize seedlings(Day 8) A: Maize physiological plant height; B: maize root length; C: plant fresh weight. CK: Sterile and salt-free group; BK: the salt-free group with strain CNY01; CN: sterile salt group; BN: the salt group with strain CNY01. *P< 0.05 and **P < 0.01, the same below
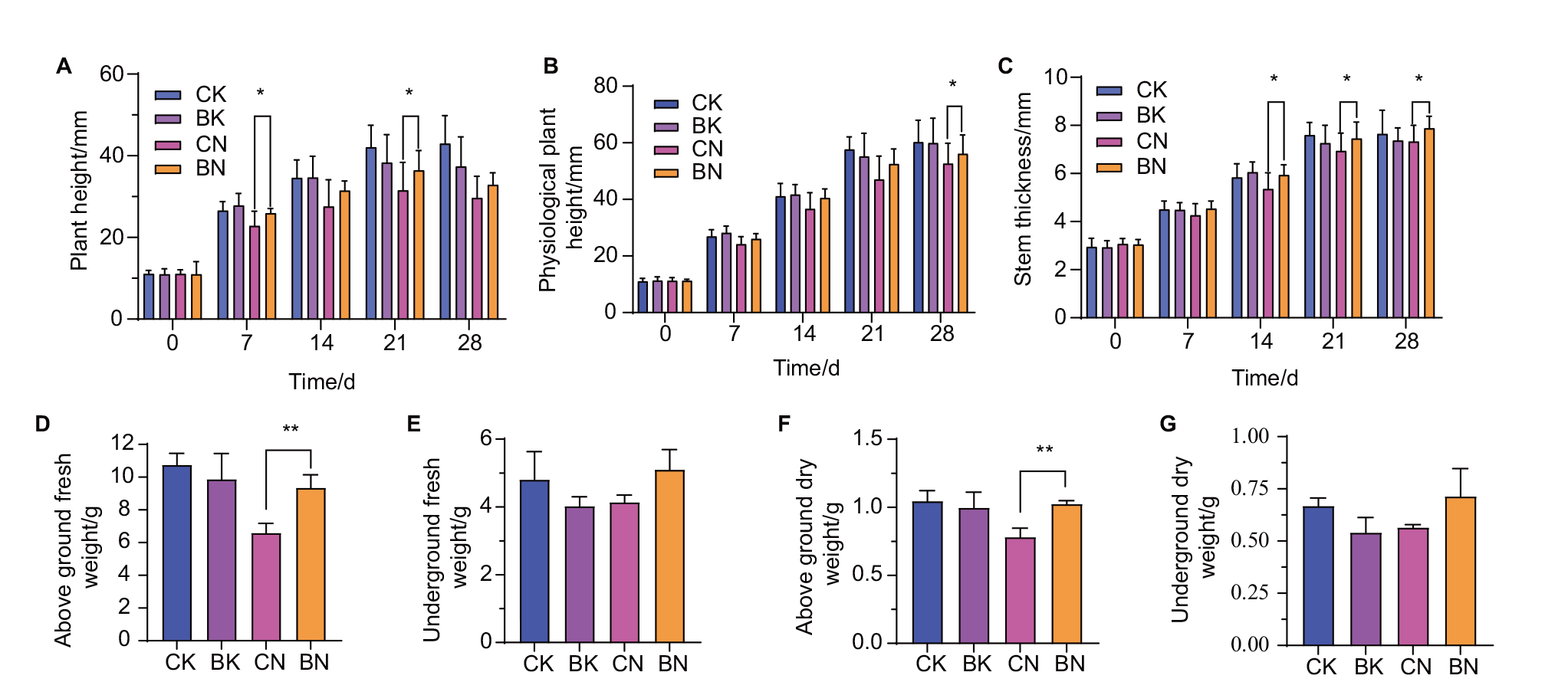
Fig. 6 Analysis of agronomic traits of maize seedlings in different periods under potted conditions A: Plant height; B: physiological plant height; C: stem thick; D: aboveground fresh weight(28 d); E: underground fresh weight(28 d); F: aboveground dry weight(28 d);G: underground dry weight(28 d)
| Genomic information | CNY01 | GQJK17 | BA59 | PENSV20 | 1942 | SRCM101359 | NS2 | NX-12 | MBLB1156 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Genome Size/Mb | 4.14 | 4.33 | 4.26 | 4.15 | 4.17 | 4.18 | 4.26 | 4.16 | 4.15 | |
| G+C content/% | 43.50 | 43.30 | 43.10 | 43.50 | 43.20 | 43.30 | 43.30 | 43.30 | 43.40 | |
| CDSs(with protein) | 3950 | 4045 | 3889 | 3932 | 3995 | 3923 | 4018 | 3946 | 3870 | |
| Pseudo genes number | 146 | 161 | 383 | 157 | 152 | 157 | 159 | 158 | 232 | |
| rRNA genes number | 24 | 24 | 26 | 24 | 21 | 24 | 24 | 24 | 24 | |
| tRNA genes number | 82 | 84 | 84 | 83 | 75 | 82 | 82 | 82 | 82 | |
| The source of isolation | Soil (China) | Rhizophere (China) | Cotton root (China) | Soil (Canada) | - | Food (South Korea) | - | Cotton rhizosphere soil | Fermented to fu |
Table 1 General genome features of B. atrophaeus CNY01, GQJK17, BA59, PENSV20, 1942, SRCM101359, NS2, NX-12, and MBLB1156
| Genomic information | CNY01 | GQJK17 | BA59 | PENSV20 | 1942 | SRCM101359 | NS2 | NX-12 | MBLB1156 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Genome Size/Mb | 4.14 | 4.33 | 4.26 | 4.15 | 4.17 | 4.18 | 4.26 | 4.16 | 4.15 | |
| G+C content/% | 43.50 | 43.30 | 43.10 | 43.50 | 43.20 | 43.30 | 43.30 | 43.30 | 43.40 | |
| CDSs(with protein) | 3950 | 4045 | 3889 | 3932 | 3995 | 3923 | 4018 | 3946 | 3870 | |
| Pseudo genes number | 146 | 161 | 383 | 157 | 152 | 157 | 159 | 158 | 232 | |
| rRNA genes number | 24 | 24 | 26 | 24 | 21 | 24 | 24 | 24 | 24 | |
| tRNA genes number | 82 | 84 | 84 | 83 | 75 | 82 | 82 | 82 | 82 | |
| The source of isolation | Soil (China) | Rhizophere (China) | Cotton root (China) | Soil (Canada) | - | Food (South Korea) | - | Cotton rhizosphere soil | Fermented to fu |
| Name of strain | Bacillus haynesii | Bacillus sonorensis | Bacillus siamensis | Bacillus licheniformis | Bacillus atrophaeus | Bacillus paralicheniformis | CNY01 |
|---|---|---|---|---|---|---|---|
| Bacillus haynesii | * | 81.18 | 71.86 | 95.14 | 72.29 | 95.14 | 72.23 |
| Bacillus sonorensis | 80.85 | * | 71.84 | 80.97 | 72.47 | 80.72 | 72.38 |
| Bacillus siamensis | 72.04 | 71.92 | * | 71.93 | 76.64 | 71.84 | 76.73 |
| Bacillus licheniformis | 95.40 | 81.42 | 71.94 | * | 72.45 | 94.38 | 72.43 |
| Bacillus atrophaeus | 72.28 | 72.57 | 76.66 | 72.30 | * | 72.32 | 98.06 |
| Bacillus paralicheniformis | 95.03 | 81.00 | 71.98 | 94.02 | 72.25 | * | 72.13 |
| CNY01 | 72.32 | 72.63 | 76.89 | 72.33 | 98.36 | 72.34 | * |
Table 2 ANI contrast test of strain CNY01
| Name of strain | Bacillus haynesii | Bacillus sonorensis | Bacillus siamensis | Bacillus licheniformis | Bacillus atrophaeus | Bacillus paralicheniformis | CNY01 |
|---|---|---|---|---|---|---|---|
| Bacillus haynesii | * | 81.18 | 71.86 | 95.14 | 72.29 | 95.14 | 72.23 |
| Bacillus sonorensis | 80.85 | * | 71.84 | 80.97 | 72.47 | 80.72 | 72.38 |
| Bacillus siamensis | 72.04 | 71.92 | * | 71.93 | 76.64 | 71.84 | 76.73 |
| Bacillus licheniformis | 95.40 | 81.42 | 71.94 | * | 72.45 | 94.38 | 72.43 |
| Bacillus atrophaeus | 72.28 | 72.57 | 76.66 | 72.30 | * | 72.32 | 98.06 |
| Bacillus paralicheniformis | 95.03 | 81.00 | 71.98 | 94.02 | 72.25 | * | 72.13 |
| CNY01 | 72.32 | 72.63 | 76.89 | 72.33 | 98.36 | 72.34 | * |
| 位置Location | 长度Length/bp | 基因Gene | 产物Product |
|---|---|---|---|
| KCX77_05795 | 453 | nhaC | Na+/H+ antiporter NhaC |
| KCX77_09890 | 429 | / | Na+/H+ antiporter NhaC family protein |
| KCX77_12210 | 468 | nhaC | Na+/H+ antiporter NhaC |
| KCX77_15895 | 806 | / | Na+/H+ antiporter subunit A |
| KCX77_15910 | 493 | / | Na+/H+ antiporter subunit D |
| KCX77_15915 | 158 | / | Na+/H+ antiporter subunit E |
| KCX77_15925 | 124 | / | Na+/H+ antiporter subunit G |
| KCX77_16910 | 671 | / | Na+/H+ antiporter |
| KCX77_01700 | 418 | opuAA | Glycine/proline betaine ABC transporter |
| KCX77_01705 | 282 | opuAB | Glycine/proline betaine ABC transporter permease subunit OpuAB |
| KCX77_01805 | 303 | / | Proline dehydrogenase |
| KCX77_01815 | 504 | putP | Sodium/proline symporter PutP |
| KCX77_04225 | 492 | putP | Sodium/proline symporter PutP |
| KCX77_09365 | 564 | proS | Proline--tRNA ligase |
| KCX77_16595 | 302 | / | Proline dehydrogenase |
| KCX77_17080 | 381 | / | Betaine/proline/choline family ABC transporter ATP-binding protein |
| KCX77_00320 | 205 | / | General stress protein Ctc |
| KCX77_04635 | 115 | / | General stress protein |
| KCX77_15790 | 130 | yugI | General stress protein 13 |
| KCX77_19470 | 547 | katX | Catalase KatX |
| KCX77_02490 | 273 | / | Manganese catalase family protein |
| KCX77_05335 | 483 | katA | Catalase KatA |
| KCX77_12565 | 285 | / | Manganese catalase family protein |
| KCX77_08110 | 302 | / | Manganese catalase family protein |
| KCX77_19655 | 685 | / | Catalase |
| KCX77_07800 | 449 | / | TrkH family potassium uptake protein |
| KCX77_08320 | 221 | ktrC | Ktr system potassium transporter KtrC |
| KCX77_15595 | 222 | / | TrkA family potassium uptake protein |
| KCX77_15600 | 445 | / | TrkH family potassium uptake protein |
| KCX77_15755 | 328 | / | Potassium channel family protein |
| KCX77_19400 | 221 | / | TrkA family potassium uptake protein |
| KCX77_17200 | 394 | / | Phosphoglycerate kinase |
| KCX77_14540 | 585 | pyk | Pyruvate kinase |
Table 3 Analysis of some genes related to salt tolerance and growth promotion in strain CNY01
| 位置Location | 长度Length/bp | 基因Gene | 产物Product |
|---|---|---|---|
| KCX77_05795 | 453 | nhaC | Na+/H+ antiporter NhaC |
| KCX77_09890 | 429 | / | Na+/H+ antiporter NhaC family protein |
| KCX77_12210 | 468 | nhaC | Na+/H+ antiporter NhaC |
| KCX77_15895 | 806 | / | Na+/H+ antiporter subunit A |
| KCX77_15910 | 493 | / | Na+/H+ antiporter subunit D |
| KCX77_15915 | 158 | / | Na+/H+ antiporter subunit E |
| KCX77_15925 | 124 | / | Na+/H+ antiporter subunit G |
| KCX77_16910 | 671 | / | Na+/H+ antiporter |
| KCX77_01700 | 418 | opuAA | Glycine/proline betaine ABC transporter |
| KCX77_01705 | 282 | opuAB | Glycine/proline betaine ABC transporter permease subunit OpuAB |
| KCX77_01805 | 303 | / | Proline dehydrogenase |
| KCX77_01815 | 504 | putP | Sodium/proline symporter PutP |
| KCX77_04225 | 492 | putP | Sodium/proline symporter PutP |
| KCX77_09365 | 564 | proS | Proline--tRNA ligase |
| KCX77_16595 | 302 | / | Proline dehydrogenase |
| KCX77_17080 | 381 | / | Betaine/proline/choline family ABC transporter ATP-binding protein |
| KCX77_00320 | 205 | / | General stress protein Ctc |
| KCX77_04635 | 115 | / | General stress protein |
| KCX77_15790 | 130 | yugI | General stress protein 13 |
| KCX77_19470 | 547 | katX | Catalase KatX |
| KCX77_02490 | 273 | / | Manganese catalase family protein |
| KCX77_05335 | 483 | katA | Catalase KatA |
| KCX77_12565 | 285 | / | Manganese catalase family protein |
| KCX77_08110 | 302 | / | Manganese catalase family protein |
| KCX77_19655 | 685 | / | Catalase |
| KCX77_07800 | 449 | / | TrkH family potassium uptake protein |
| KCX77_08320 | 221 | ktrC | Ktr system potassium transporter KtrC |
| KCX77_15595 | 222 | / | TrkA family potassium uptake protein |
| KCX77_15600 | 445 | / | TrkH family potassium uptake protein |
| KCX77_15755 | 328 | / | Potassium channel family protein |
| KCX77_19400 | 221 | / | TrkA family potassium uptake protein |
| KCX77_17200 | 394 | / | Phosphoglycerate kinase |
| KCX77_14540 | 585 | pyk | Pyruvate kinase |
| [1] | 王佳丽, 黄贤金, 钟太洋, 等. 盐碱地可持续利用研究综述[J]. 地理学报, 2011, 66(5): 673-684. |
|
Wang JL, Huang XJ, Zhong TY, et al. Review on sustainable utilization of salt-affected land[J]. Acta Geogr Sin, 2011, 66(5): 673-684.
doi: 10.11821/xb201105010 |
|
| [2] | 魏博娴. 中国盐碱土的分布与成因分析[J]. 水土保持应用技术, 2012(6): 27-28. |
| Wei BX. Distribution and cause analysis of saline-alkali soil in China[J]. Technol Soil Water Conserv, 2012(6): 27-28. | |
| [3] | 赵娇. 基于菌群改善促进盐碱地耐受性植物生长的研究[D]. 济南: 山东大学, 2020. |
| Zhao J. Study on the improvement of flora to promote thegrowth of tolerant plants in saline-alkali land[D]. Jinan: Shandong University, 2020. | |
| [4] | 王文飞. 萎缩芽孢杆菌WU-9与辣椒互作缓解盐胁迫机制及菌株相关功能基因解析[D]. 石河子: 石河子大学, 2022. |
| Wang WF. Mechanism of relieving salt stress by interaction between Bacillus Atrophyta WU-9 and pepper and analysis of related functional genes of the strain[D]. Shihezi: Shihezi University, 2022. | |
| [5] | 郭英. 生防芽孢杆菌对作物耐盐性的影响及其抗盐机理的研究[D]. 济南: 山东师范大学, 2009. |
| Guo Y. Effects and mechanism of biocontrol strain of Bacillus subtilis on salt tolerance of plants[D]. Jinan: Shandong Normal University, 2009. | |
| [6] | 庞亚琴, 任彩婷, 徐秋曼. 解淀粉芽孢杆菌HM618对镉胁迫下小麦幼苗生长的影响[J]. 天津师范大学学报: 自然科学版, 2018, 38(4): 55-59. |
| Pang YQ, Ren CT, Xu QM. Effects of Bacillus amyloliquefaciens HM618 on the growth of wheat seedlings under cadmium stress[J]. J Tianjin Norm Univ Nat Sci Ed, 2018, 38(4): 55-59. | |
| [7] |
Sella SRBR, Vandenberghe LPS, Soccol CR. Bacillus atrophaeus: main characteristics and biotechnological applications - a review[J]. Crit Rev Biotechnol, 2015, 35(4): 533-545.
doi: 10.3109/07388551.2014.922915 pmid: 24963702 |
| [8] | Ma JJ, Wang CQ, Wang HD, et al. Analysis of the complete genome sequence of Bacillus atrophaeus GQJK17 reveals its biocontrol characteristics as a plant growth-promoting rhizobacterium[J]. Biomed Res Int, 2018, 2018: 9473542. |
| [9] |
Gordon RE, Smith NR. Aerobic sporeforming bacteria capable of growth at high temperatures[J]. J Bacteriol, 1949, 58(3): 327-341.
doi: 10.1128/jb.58.3.327-341.1949 pmid: 16561790 |
| [10] | Nakamura LK. Taxonomic relationship of black-pigmented Bacillus subtilis strains and a proposal for Bacillus atrophaeus sp. nov.[J]. International Journal of Systematic and Evolutionary Microbiology, 1989, 39(3):295-300. |
| [11] | Hou YL, Zeng WZ, Ao C, et al. Bacillus atrophaeus WZYH01 and Planococcus soli WZYH02 improve salt tolerance of maize(Zea mays L.) in saline soil[J]. Front Plant Sci, 2022, 13: 891372. |
| [12] | 韦廷舟, 文怡, 王超, 等. 一株产IAA芽孢杆菌ST37对油菜的耐盐促生作用[J]. 江苏农业科学, 2023, 51(6): 210-215. |
| Wei TZ, Wen Y, Wang C, et al. Effect of an IAA-producing Bacillus ST37 on salt tolerance and growth promotion of rape[J]. Jiangsu Agric Sci, 2023, 51(6): 210-215. | |
| [13] | 郇惠杰, 钟泓波, 雷芬芬, 等. 产蛋白酶海洋细菌的筛选、鉴定及发酵培养基的研究[J]. 食品工业科技, 2013, 34(24): 181-185. |
| Huan HJ, Zhong HB, Lei FF, et al. Study on isolation and identification of protease-producing marine bacteria and optimization of fermentation medium[J]. Sci Technol Food Ind, 2013, 34(24): 181-185. | |
| [14] | 张祥胜. 发酵液有效磷含量测定方法研究[J]. 湖州职业技术学院学报, 2008, 6(3): 1-3. |
| Zhang XS. A study of factors affecting the determined value by Mo-Sn-vc method of organic phosphobacteria[J]. J Huzhou Vocat Technol Coll, 2008, 6(3): 1-3. | |
| [15] | 李浩. 玉米、黄瓜根际促生菌组合优化及基因组测序[D]. 泰安: 山东农业大学, 2019. |
| Li H. Optimization of rhizosphere growth-promoting bacteria combination and genome sequencing of maize and cucumber[D]. Tai'an: Shandong Agricultural University, 2019. | |
| [16] | 马锦锦. 枸杞根际促生细菌筛选、培养基优化及基因组测序[D]. 泰安: 山东农业大学, 2018. |
| Ma JJ. Screening, Medium optimization and genome sequencing of PGPR from the rhizosphere of Lycium barbarum L.[D]. Tai'an: Shandong Agricultural University, 2018. | |
| [17] | 侯贞. 烟草根际促生细菌的筛选鉴定及发酵培养基优化[D]. 泰安: 山东农业大学, 2015. |
| Hou Z. Screening and identification of tobacco rhizosphere growth-promoting bacteria and optimization of fermentation medium[D]. Tai'an: Shandong Agricultural University, 2015. | |
| [18] | 杨亚男. 番茄根际促生菌的筛选及其培养基优化[D]. 泰安: 山东农业大学, 2017. |
| Yang YN. Screening of tomato rhizosphere growth-promoting bacteria and optimization of its culture medium[D]. Tai'an: Shandong Agricultural University, 2017. | |
| [19] |
Schubert M, Lindgreen S, Orlando L. AdapterRemoval v2: rapid adapter trimming, identification, and read merging[J]. BMC Res Notes, 2016, 9: 88.
doi: 10.1186/s13104-016-1900-2 pmid: 26868221 |
| [20] | Luo RB, Liu BH, Xie YL, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler[J]. GigaScience, 2012, 1(1): 18. |
| [21] |
Chin CS, Peluso P, Sedlazeck FJ, et al. Phased diploid genome assembly with single-molecule real-time sequencing[J]. Nat Methods, 2016, 13(12): 1050-1054.
doi: 10.1038/NMETH.4035 |
| [22] | Koren S, Walenz BP, Berlin K, et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation[J]. Genome Res, 2017, 27(5): 722-736. |
| [23] | Walker BJ, Abeel T, Shea T, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement[J]. PLoS One, 2014, 9(11): e112963. |
| [24] | 王欣. 植物根际促生细菌CNY01和BY2G20的耐盐功能及其对玉米的抗盐促生研究[D]. 泰安: 山东农业大学, 2021. |
| Wang X, The Salt Tolerance Function of Plant Growth-promoting Rhizobacteria CNY01 and BY2G20 and Their Anti-salt Promotion for Corn Growth[D]. Tai'an: Shandong Agricultural University, 2021. | |
| [25] | 索雲凯, 刘丽红, 张雷, 等. 解钾菌解钾作用研究进展[J]. 当代化工, 2021, 50(4): 924-929. |
| Suo YK, Liu LH, Zhang L, et al. Research progress of potassium solubilization by potassium solubilizing bacteria[J]. Contemp Chem Ind, 2021, 50(4): 924-929. | |
| [26] | Ayaz M, Ali Q, Farzand A, et al. Nematicidal volatiles from Bacillus atrophaeus GBSC56 promote growth and stimulate induced systemic resistance in tomato against Meloidogyne incognita[J]. Int J Mol Sci, 2021, 22(9): 5049. |
| [27] | 刘思靖. 萎缩芽孢杆菌次级代谢产物的研究进展[J]. 现代化工, 2019, 39(12): 48-51. |
| Liu SJ. Advances in secondary metabolites from Bacillus atrophaeus[J]. Mod Chem Ind, 2019, 39(12): 48-51. | |
| [28] |
戴宝, 扈进冬, 尹姗姗, 等. 响应面法优化萎缩芽孢杆菌BsR05发酵培养基[J]. 山东科学, 2017, 30(4): 31-37.
doi: 10.3976/j.issn.1002-4026.2017.04.006 |
| Dai B, Hu JD, Yin SS, et al. Optimization of fermentation medium composition for Bacillus atrophaeus BsR05 by response surface method[J]. Shandong Sci, 2017, 30(4): 31-37. | |
| [29] | 王敏. 萎缩芽孢杆菌XW2对苹果树腐烂病的防效评价[D]. 乌鲁木齐: 新疆农业大学, 2020. |
| Wang M. Evaluation on control effect of Bacillus atrophyta XW2 on apple tree rot[D]. Urumqi: Xinjiang Agricultural University, 2020. | |
| [30] |
胡玉婕, 朱秀玲, 丁延芹, 等. 芽孢杆菌的耐盐促生机制研究进展[J]. 生物技术通报, 2020, 36(9): 64-74.
doi: 10.13560/j.cnki.biotech.bull.1985.2020-0746 |
| Hu YJ, Zhu XL, Ding YQ, et al. Research progress on salt tolerance and growth-promoting mechanism of Bacillus[J]. Biotechnol Bull, 2020, 36(9): 64-74. | |
| [31] | Karim R, Bouchra B, Fatima G, et al. Plant NHX antiporters: from function to biotechnological application, with case study[J]. Curr Protein Pept Sci, 2021, 22(1): 60-73. |
| [32] | 赵东晓, 董亚茹, 孙景诗, 等. NaCl胁迫对胡麻种子萌发、幼苗生长及Na+/H+逆向转运蛋白基因表达的影响[J]. 山东农业科学, 2020, 52(7): 40-45. |
| Zhao DX, Dong YR, Sun JS, et al. Effects of NaCl stress on seed germination, seedling growth and Na+/H+ antiporter protein gene expression of Linum usitatissimum L[J]. Shandong Agric Sci, 2020, 52(7): 40-45. | |
| [33] |
张傲洁, 李青云, 宋文红, 等. 基于苯酚降解的粪产碱杆菌Alcaligenes faecalis JF101的全基因组分析[J]. 生物技术通报, 2023, 39(10): 292-303.
doi: 10.13560/j.cnki.biotech.bull.1985.2023-0281 |
| Zhang AJ, Li QY, Song WH, et al. Whole genome sequencing analysis of a phenol-degrading strain Alcaligenes faecalis JF101[J]. Biotechnol Bull, 2023, 39(10): 292-303. |
| [1] | WANG Jia-wei, LI Chen, LIU Jian-li, ZHOU Shi-jie, YI Jia-min, YANG Jin-yuan, KANG Peng. Effects of Endophytic Fungal Inoculation on the Seedling Growth of Silage Maize [J]. Biotechnology Bulletin, 2024, 40(4): 189-202. |
| [2] | HU Yi-wa, CHEN Lu. Research Advance and Applications in Maize Wild Relatives Genomes [J]. Biotechnology Bulletin, 2024, 40(3): 14-24. |
| [3] | CHANG Lu-yin, WANG Zhong-hua, LI Feng-min, GAO Zi-yuan, ZHANG Hui-hong, WANG Yi, LI Fang, HAN Yan-lai, JIANG Ying. Screening Multi-functional Rhizobacteria from Maize Rhizosphere and Their Ehancing Effects on Winter Wheat-Summer Maize Rotation System [J]. Biotechnology Bulletin, 2024, 40(1): 231-242. |
| [4] | WANG Bao-bao, WANG Hai-yang. Molecular Design of Ideal Plant Architecture for High-density Tolerance of Maize Plant [J]. Biotechnology Bulletin, 2023, 39(8): 11-30. |
| [5] | ZHANG Dao-lei, GAN Yu-jun, LE Liang, PU Li. Epigenetic Regulation of Yield-related Traits in Maize and Epibreeding [J]. Biotechnology Bulletin, 2023, 39(8): 31-42. |
| [6] | LENG Yan, MA Xiao-wei, CHEN Guang, REN He, LI Xiang. High-yield Contests in Maize Facilitate the Vitalization of China’s Seed Industry [J]. Biotechnology Bulletin, 2023, 39(8): 4-10. |
| [7] | WANG Tian-yi, WANG Rong-huan, WANG Xia-qing, ZHANG Ru-yang, XU Rui-bin, JIAO Yan-yan, SUN Xuan, WANG Ji-dong, SONG Wei, ZHAO Jiu-ran. Research in Maize Dwarf Genes and Dwarf Breeding [J]. Biotechnology Bulletin, 2023, 39(8): 43-51. |
| [8] | LIU Yue-e, XU Tian-jun, CAI Wan-tao, LYU Tian-fang, ZHANG Yong, XUE Hong-he, WANG Rong-huan, ZHAO Jiu-ran. Current Status and Prospects of Maize Super High Yield Research in China [J]. Biotechnology Bulletin, 2023, 39(8): 52-61. |
| [9] | ZHU Shao-xi, JIN Zhao-yang, GE Jian-rong, WANG Rui, WANG Feng-ge, LU Yun-cai. High-throughput Specific Detection Methods for Transgenic Maize Based on the KASP Platform [J]. Biotechnology Bulletin, 2023, 39(6): 133-140. |
| [10] | CHEN Nan-nan, WANG Chun-lai, JIANG Zhen-zhong, JIAO Peng, GUAN Shu-yan, MA Yi-yong. Genetic Transformation and Chilling Resistance Analysis of Maize ZmDHN15 Gene in Tobacco [J]. Biotechnology Bulletin, 2023, 39(4): 259-267. |
| [11] | LI Sheng-yan, LI Xiang-yin, LI Peng-cheng, ZHANG Ming-jun, ZHANG Jie, LANG Zhi-hong. Identification of Target Traits and Genetic Stability of Transgenic Maize 2HVB5 [J]. Biotechnology Bulletin, 2023, 39(1): 21-30. |
| [12] | LI Dong-yang, XIAO Bing, WANG Chen-yao, YANG Xian-ming, LIANG Jin-gang, WU Kong-ming. Spatio-temporal Expression of Cry1Ab/Cry2Aj Insecticidal Protein in Genetically Modified Maize Ruifeng 125 with Stacked Insect and Herbicide Resistance Traits [J]. Biotechnology Bulletin, 2023, 39(1): 31-39. |
| [13] | LI Peng-cheng, ZHANG Ming-jun, WANG Yin-xiao, LI Xiang-yin, LI Sheng-yan, LANG Zhi-hong. Insect Resistance Identification and Agronomy Traits Analysis of Transgenic Maize HGK60 with Different Genetic Backgrounds [J]. Biotechnology Bulletin, 2023, 39(1): 40-47. |
| [14] | JIN Yun-qian, WANG Bin, GUO Shu-lei, ZHAO Lin-xi, HAN Zan-ping. Research Progress in Gibberellin Regulation on Maize Seed Vigor [J]. Biotechnology Bulletin, 2023, 39(1): 84-94. |
| [15] | ZHU Jing, YU Cun. Effects of Trichoderma longibrachiatum on Maize Growth,Soil Fertility and Rhizosphere Microorganism [J]. Biotechnology Bulletin, 2022, 38(4): 230-241. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||