








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (9): 104-112.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0243
Previous Articles Next Articles
SONG Bing-fang( ), LIU Ning, CHENG Xin-yan, XU Xiao-bin, TIAN Wen-mao, GAO Yue, BI Yang, WANG Yi(
), LIU Ning, CHENG Xin-yan, XU Xiao-bin, TIAN Wen-mao, GAO Yue, BI Yang, WANG Yi( )
)
Received:2024-03-12
Online:2024-09-26
Published:2024-07-31
Contact:
WANG Yi
E-mail:3132897396@qq.com;wangyi@gsau.edu.cn
SONG Bing-fang, LIU Ning, CHENG Xin-yan, XU Xiao-bin, TIAN Wen-mao, GAO Yue, BI Yang, WANG Yi. Identification of Potato G6PDH Gene Family and Its Expression Analysis in Damaged Tubers[J]. Biotechnology Bulletin, 2024, 40(9): 104-112.
| 基因名称 Gene name | 基因ID Gene ID | 引物序列 Primer sequence(5'-3') |
|---|---|---|
| StG6PDH1 | Soltu.DM.01G040700 | F: GCTTTCACCAGTATCGCTATCA |
| R: CGGACAACAACGGTAGGTAAT | ||
| StG6PDH2 | Soltu.DM.02G033900 | F: CAAGAAACCTGGGCTTGAAATG |
| R: CGTTCATAAGCCTCTGGAATGA | ||
| StG6PDH3 | Soltu.DM.05G012290 | F: ATACCAGGAGTGCGGAAATAAG |
| R: CGTCAGGTTGAACACGGATAA | ||
| StG6PDH4 | Soltu.DM.07G015360 | F: GCTGACCAGATCCCTAAAGAAG |
| R: GACAAGATTCGAGAACCGAAGA | ||
| StEF1α | Soltu.DM.06G005580 | F: ATTGATGCCCCTGGTCACAG |
| R: CATGTTCACGGGTCTGACCA |
Table 1 Primers’ sequences for RT-qPCR
| 基因名称 Gene name | 基因ID Gene ID | 引物序列 Primer sequence(5'-3') |
|---|---|---|
| StG6PDH1 | Soltu.DM.01G040700 | F: GCTTTCACCAGTATCGCTATCA |
| R: CGGACAACAACGGTAGGTAAT | ||
| StG6PDH2 | Soltu.DM.02G033900 | F: CAAGAAACCTGGGCTTGAAATG |
| R: CGTTCATAAGCCTCTGGAATGA | ||
| StG6PDH3 | Soltu.DM.05G012290 | F: ATACCAGGAGTGCGGAAATAAG |
| R: CGTCAGGTTGAACACGGATAA | ||
| StG6PDH4 | Soltu.DM.07G015360 | F: GCTGACCAGATCCCTAAAGAAG |
| R: GACAAGATTCGAGAACCGAAGA | ||
| StEF1α | Soltu.DM.06G005580 | F: ATTGATGCCCCTGGTCACAG |
| R: CATGTTCACGGGTCTGACCA |
| 基因名称 Gene name | 基因ID Gene ID | 染色体位置 Chromosome location | 亚细胞定位 Subcellular location |
|---|---|---|---|
| StG6PDH1 | Soltu.DM.01G040700 | chr.01 | 叶绿体Chloroplast |
| StG6PDH2 | Soltu.DM.02G033900 | chr.02 | 细胞质Cytoplasmic |
| StG6PDH3 | Soltu.DM.05G012290 | chr.05 | 叶绿体Chloroplast |
| StG6PDH4 | Soltu.DM.07G015360 | chr.07 | 叶绿体Chloroplast |
Table 2 Prediction of subcellular localization of G6PDH protein in potato
| 基因名称 Gene name | 基因ID Gene ID | 染色体位置 Chromosome location | 亚细胞定位 Subcellular location |
|---|---|---|---|
| StG6PDH1 | Soltu.DM.01G040700 | chr.01 | 叶绿体Chloroplast |
| StG6PDH2 | Soltu.DM.02G033900 | chr.02 | 细胞质Cytoplasmic |
| StG6PDH3 | Soltu.DM.05G012290 | chr.05 | 叶绿体Chloroplast |
| StG6PDH4 | Soltu.DM.07G015360 | chr.07 | 叶绿体Chloroplast |
| 基因名称 Gene name | 理化性质Physicochemical property | 二级结构Secondary structure | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 氨基酸长度 Amino acid length/aa | 相对分子量 Mw/kD | 等电点 pI | 不稳定系数Instability index | α-螺旋 α-helix/% | β-转角 β-turn/% | 无规则卷曲 Random coil/% | 延伸链 Extended strand/% | |||
| StG6PDH1 | 596 | 66.65 | 5.83 | 47.31 | 36.91 | 5.87 | 40.27 | 16.95 | ||
| StG6PDH2 | 511 | 58.48 | 5.97 | 47.53 | 40.51 | 5.28 | 39.33 | 16.87 | ||
| StG6PDH3 | 582 | 66.21 | 8.57 | 39.79 | 38.14 | 6.53 | 39.87 | 17.53 | ||
| StG6PDH4 | 577 | 65.72 | 6.88 | 39.84 | 37.78 | 6.59 | 39.17 | 16.46 | ||
Table 3 Protein physicochemical properties and secondary structures of potato G6PDH gene family
| 基因名称 Gene name | 理化性质Physicochemical property | 二级结构Secondary structure | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 氨基酸长度 Amino acid length/aa | 相对分子量 Mw/kD | 等电点 pI | 不稳定系数Instability index | α-螺旋 α-helix/% | β-转角 β-turn/% | 无规则卷曲 Random coil/% | 延伸链 Extended strand/% | |||
| StG6PDH1 | 596 | 66.65 | 5.83 | 47.31 | 36.91 | 5.87 | 40.27 | 16.95 | ||
| StG6PDH2 | 511 | 58.48 | 5.97 | 47.53 | 40.51 | 5.28 | 39.33 | 16.87 | ||
| StG6PDH3 | 582 | 66.21 | 8.57 | 39.79 | 38.14 | 6.53 | 39.87 | 17.53 | ||
| StG6PDH4 | 577 | 65.72 | 6.88 | 39.84 | 37.78 | 6.59 | 39.17 | 16.46 | ||
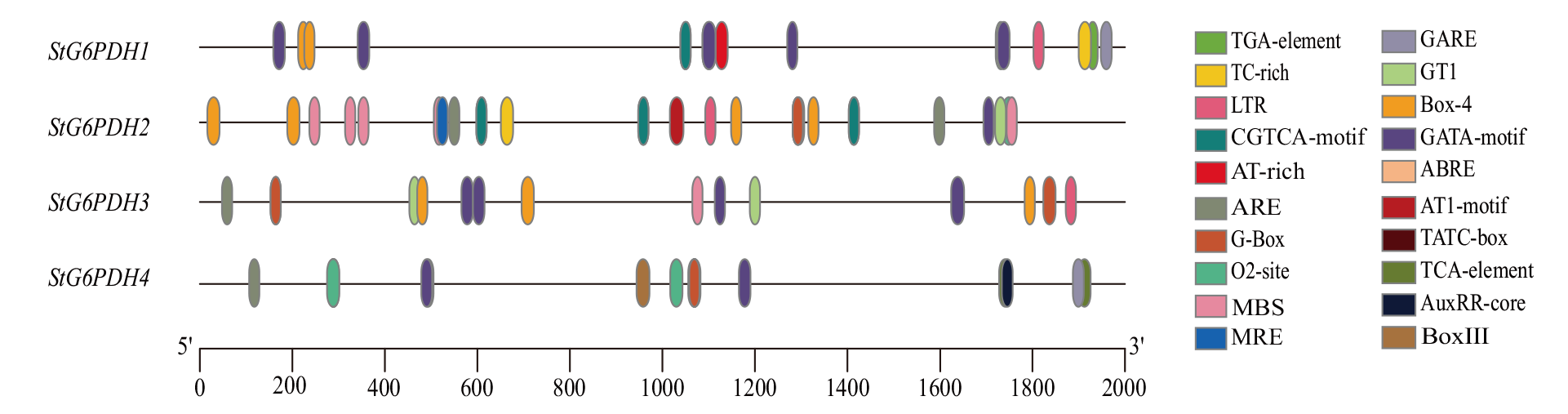
Fig. 4 Cis-acting elements of potato G6PDH gene family Light responsiveness: GATA-motif, GT1, Box-4, MRE, G-Box, AT1-motif. Low-temperature responsiveness: LTR. Anaerobic induction: ARE. Gibberellin responsiveness: GARE, TATC-box. Auxin responsiveness: AuxRE-core, TGA-element. Abscisic acid responsiveness: ABRE. Me-JA responsiveness: CGTCA-motif. Salicylic acid responsiveness: TCA-element. Drought-induced response element: MBS. Zein metabolism regulation: O2-site. Defense and stress responsiveness: TC-rich. Other: Box III, AT-rich
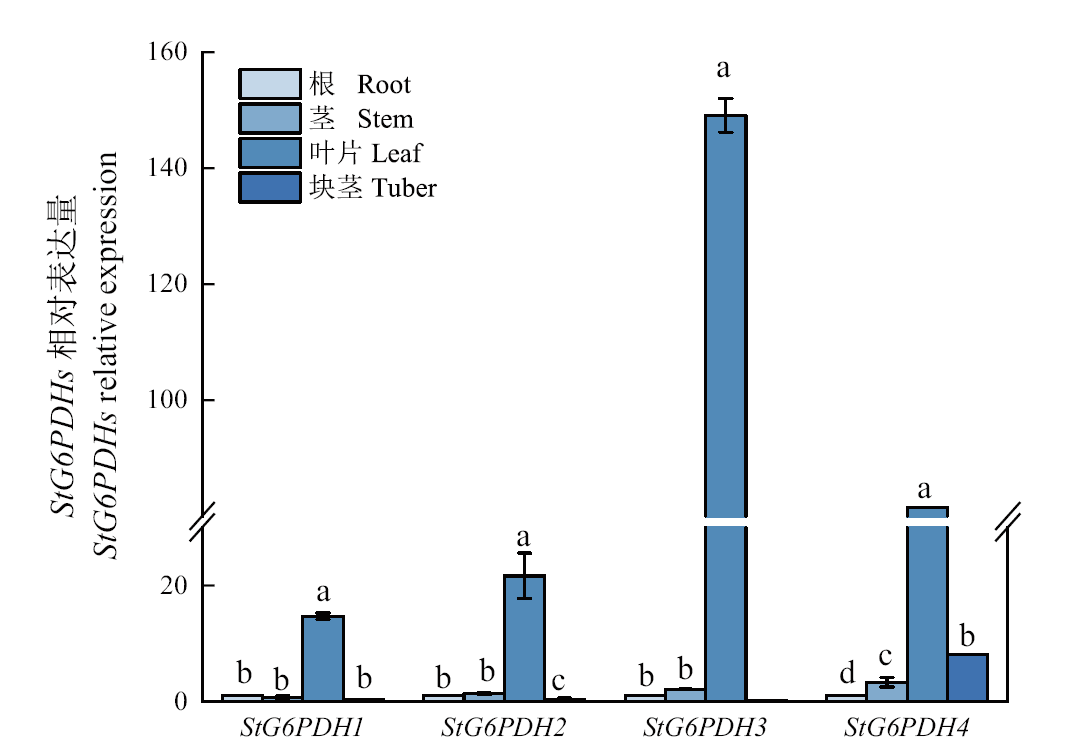
Fig. 5 Relative expressions of G6PDH in different organs of potato Different lowercase letters in the same gene indicate significant difference(P<0.05)
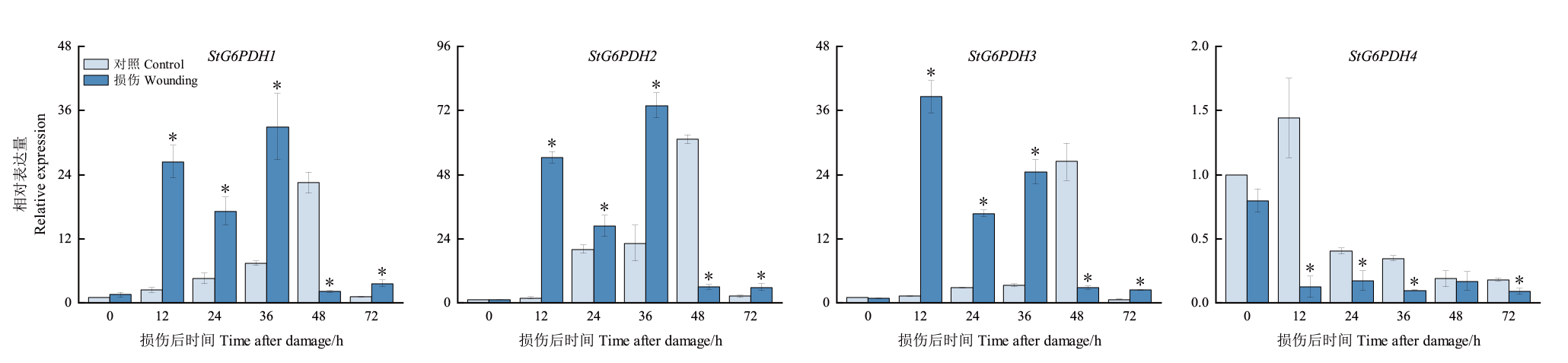
Fig. 6 Relative expression of StG6PDH1s in damaged potato tubers * indicates a significant difference between the control group and the treatment group(P<0.05)
| [1] |
Beumer K, Stemerding D. A breeding consortium to realize the potential of hybrid diploid potato for food security[J]. Nat Plants, 2021, 7(12): 1530-1532.
doi: 10.1038/s41477-021-01035-4 pmid: 34815537 |
| [2] | Hu WZ, Guan YG, Ji YR, et al. Effect of cutting styles on quality, antioxidant activity, membrane lipid peroxidation, and browning in fresh-cut potatoes[J]. Food Biosci, 2021, 44: 101435. |
| [3] | París R, Lamattina L, Casalongué CA. Nitric oxide promotes the wound-healing response of potato leaflets[J]. Plant Physiol Biochem, 2007, 45(1): 80-86. |
| [4] |
姜红, 王毅, 毕阳. 马铃薯块茎的愈伤过程、机制和影响因素[J]. 园艺学报, 2019, 46(9): 1842-1852.
doi: 10.16420/j.issn.0513-353x.2019-0358 |
| Jiang H, Wang Y, Bi Y. Healing processes, mechanisms and factors affecting potato tubers[J]. Journal of Horticulture, 2019, 46(9): 1842-1852. | |
| [5] | Bussell JD, Keech O, Fenske R, et al. Requirement for the plastidial oxidative pentose phosphate pathway for nitrate assimilation in Arabidopsis[J]. Plant J, 2013, 75(4): 578-591. |
| [6] |
Corpas FJ, González-Gordo S, Palma JM. Nitric oxide and hydrogen sulfide modulate the NADPH-generating enzymatic system in higher plants[J]. J Exp Bot, 2021, 72(3): 830-847.
doi: 10.1093/jxb/eraa440 pmid: 32945878 |
| [7] | Chen PH, Tjong WY, Yang HC, et al. Glucose-6-phosphate dehydrogenase, redox homeostasis and embryogenesis[J]. Int J Mol Sci, 2022, 23(4): 2017. |
| [8] | Landi S, Nurcato R, De Lillo A, et al. Glucose-6-phosphate dehydrogenase plays a central role in the response of tomato(Solanum lycopersicum)plants to short and long-term drought[J]. Plant Physiol Biochem, 2016, 105: 79-89. |
| [9] | Li X, Cai Q, Yu T, et al. ZmG6PDH1 in glucose-6-phosphate dehydrogenase family enhances cold stress tolerance in maize[J]. Front Plant Sci, 2023, 14: 1116237. |
| [10] | Zhao Y, Cui YF, Huang SY, et al. Genome-wide analysis of the glucose-6-phosphate dehydrogenase family in soybean and functional identification of GmG6PDH2 involvement in salt stress[J]. Front Plant Sci, 2020, 11: 214. |
| [11] | Li XA, Li BR, Min DD, et al. Transcriptomic analysis reveals key genes associated with the biosynthesis regulation of phenolics in fresh-cut pitaya fruit(Hylocereus undatus)[J]. Postharvest Biol Technol, 2021, 181: 111684. |
| [12] | Chintha P, Sarkar D, Ramakrishna R, et al. Biological elicitors to enhance wound healing responses in cut potato tubers[J]. Sci Hortic, 2023, 319: 112152. |
| [13] | Wakao S, Benning C. Genome-wide analysis of glucose-6-phosphate dehydrogenases in Arabidopsis[J]. Plant J, 2005, 41(2): 243-256. |
| [14] |
Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant, 2020, 13(8): 1194-1202.
doi: S1674-2052(20)30187-8 pmid: 32585190 |
| [15] | 韩占红, 王斌, 杨瑞瑞, 等. 一氧化氮处理对马铃薯采后块茎愈伤的促进及机制[J]. 食品科学, 2020, 41(21): 222-229. |
| Han ZH, Wang B, Yang RR, et al. Effect and mechanism of postharvest nitric oxide treatment on promoting wound healing in potato tubers[J]. Food Sci, 2020, 41(21): 222-229. | |
| [16] | Esposito S. Nitrogen assimilation, abiotic stress and glucose 6-phosphate dehydrogenase: the full circle of reductants[J]. Plants, 2016, 5(2): 24. |
| [17] | Yang YT, Fu ZW, Su YC, et al. A cytosolic glucose-6-phosphate dehydrogenase gene, ScG6PDH, plays a positive role in response to various abiotic stresses in sugarcane[J]. Sci Rep, 2014, 4: 7090. |
| [18] | Hou FY, Huang J, Yu SL, et al. The 6-phosphogluconate dehydrogenase genes are responsive to abiotic stresses in rice[J]. J Integr Plant Biol, 2007, 49(5): 655-663. |
| [19] | 田宇, 彭瞰看, 宋春华, 等. 小麦G6PDH基因的生物信息学分析及其低温胁迫下苗期的表达特征[J]. 麦类作物学报, 2019, 39(6): 631-638. |
| Tian Y, Peng KK, Song CH, et al. Bioinformatics analysis of wheat G6PDH genes and their patterns in tillering node and leaf under cold stress[J]. J Triticeae Crops, 2019, 39(6): 631-638. | |
| [20] | Zhang YT, Luo MW, Cheng LJ, et al. Identification of the cytosolic glucose-6-phosphate dehydrogenase gene from strawberry involved in cold stress response[J]. Int J Mol Sci, 2020, 21(19): 7322. |
| [21] | Jiang ZR, Wang M, Nicolas M, et al. Glucose-6-phosphate dehydrogenases: the hidden players of plant physiology[J]. Int J Mol Sci, 2022, 23(24): 16128. |
| [22] | Landi S, Capasso G, Esposito S. Different G6PDH isoforms show specific roles in acclimation to cold stress at various growth stages of barley(Hordeum vulgare)and Arabidopsis thaliana[J]. Plant Physiol Biochem, 2021, 169: 190-202. |
| [23] | Whisstock JC, Lesk M. 预测的蛋白质的功能的蛋白质的顺序和结构[J]. 季度审查的生物物理学, 2003, 36(3): 307-340. |
| Whisstock JC, Lesk M. Order and structure of proteins for predicted protein function[J]. Quarterly Review of Biophysics, 2003, 36(3): 307-340. | |
| [24] | 车卓, 张沛沛, 陈涛, 等. 小麦G6PDH基因家族的鉴定与表达分析[J]. 麦类作物学报, 2023, 43(8): 947-957. |
| Che Z, Zhang PP, Chen T, et al. Identification and expression analysis of the G6PDH gene family in wheat[J]. J Triticeae Crops, 2023, 43(8): 947-957. | |
| [25] |
Preiser AL, Fisher N, Banerjee A, et al. Plastidic glucose-6-phosphate dehydrogenases are regulated to maintain activity in the light[J]. Biochem J, 2019, 476(10): 1539-1551.
doi: 10.1042/BCJ20190234 pmid: 31092702 |
| [26] | Lei DY, Lin YX, Luo MW, et al. Genome-wide investigation of G6PDH gene in strawberry: evolution and expression analysis during development and stress[J]. Int J Mol Sci, 2022, 23(9): 4728-4728. |
| [27] | Cardi M, Chibani K, Cafasso D, et al. Abscisic acid effects on activity and expression of barley(Hordeum vulgare)plastidial glucose-6-phosphate dehydrogenase[J]. J Exp Bot, 2011, 62(11): 4013-4023. |
| [28] | Tian Y, Peng KK, Bao YZ, et al. Glucose-6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase genes of winter wheat enhance the cold tolerance of transgenic Arabidopsis[J]. Plant Physiol Biochem, 2021, 161: 86-97. |
| [29] | Wang HH, Yang LD, Li Y, et al. Involvement of ABA- and H2O2-dependent cytosolic glucose-6-phosphate dehydrogenase in maintaining redox homeostasis in soybean roots under drought stress[J]. Plant Physiol Biochem, 2016, 107: 126-136. |
| [30] | Feng RJ, Wang XM, He L, et al. Identification, characterization, and stress responsiveness of glucose-6-phosphate dehydrogenase genes in highland barley[J]. Plants, 2020, 9(12): 1800. |
| [31] |
Knight JS, Emes MJ, Debnam PM. Isolation and characterisation of a full-length genomic clone encoding a plastidic glucose 6-phosphate dehydrogenase from Nicotiana tabacum[J]. Planta, 2001, 212(4): 499-507.
doi: 10.1007/s004250000419 pmid: 11525506 |
| [32] |
Weise SE, Liu T, Childs KL, et al. Transcriptional regulation of the glucose-6-phosphate/ phosphate translocator 2 is related to carbon exchange across the chloroplast envelope[J]. Front Plant Sci, 2019, 10: 827.
doi: 10.3389/fpls.2019.00827 pmid: 31316533 |
| [33] | Gao S, Zheng ZB, Huan L, et al. G6PDH activity highlights the operation of the cyclic electron flow around PSI in Physcomitrella patens during salt stress[J]. Sci Rep, 2016, 6: 21245. |
| [34] | Wei XB, Huang XL, Yang WL, et al. A chloroplast-localized glucose-6-phosphate dehydrogenase positively regulates stripe rust resistance in wheat[J]. Int J Mol Sci, 2022, 24(1): 459. |
| [35] |
Lulai EC, Suttle JC. Signals involved in tuber wound-healing[J]. Plant Signal Behav, 2009, 4(7): 620-622.
doi: 8922 pmid: 19820323 |
| [36] | Wang L, Wang WX, Shan JW, et al. A genome-wide view of the transcriptome dynamics of fresh-cut potato tubers[J]. Genes, 2023, 14(1): 181. |
| [1] | SHEN Peng, GAO Ya-Bin, DING Hong. Identification and Expression Analysis of SAT Gene Family in Potato(Solanum tuberosum L.) [J]. Biotechnology Bulletin, 2024, 40(9): 64-73. |
| [2] | WANG Chao, BAI Ru-qian, GUAN Jun-mei, LUO Ji-lin, HE Xue-jiao, CHI Shao-yi, MA Ling. Promotion of StHY5 in the Synthesis of SGAs during Tuber Turning-green of Potato [J]. Biotechnology Bulletin, 2024, 40(9): 113-122. |
| [3] | XIA Shi-xuan, GENG Ze-dong, ZHU Guang-tao, ZHANG Chun-zhi, LI Da-wei. Quick Detection of Potato Pollen Viability Based on Deep Learning [J]. Biotechnology Bulletin, 2024, 40(9): 123-130. |
| [4] | MAO Xiang-hong, LU Yao, FAN Xiang-bin, DU Pei-bing, BAI Xiao-dong. Genetic Diversity Analysis of Potato Varieties Based on SSR Fluorescent Marker Capillary Electrophoresis and Construction of Molecular Identity Card [J]. Biotechnology Bulletin, 2024, 40(9): 131-140. |
| [5] | YUAN Lan, HUANG Ya-nan, ZHANG Bei-ni, XIONG Yu-meng, WANG Hong-yang. High-throughput Sample Preparation Method for the Identification of Potato Ploidy Using Flow Cytometry [J]. Biotechnology Bulletin, 2024, 40(9): 141-147. |
| [6] | WU Hui-qin, WANG Yan-hong, LIU Han, SI Zheng, LIU Xue-qing, WANG Jing, YANG Yi, CHENG Yan. Identification and Expression Analysis of UGT Gene Family in Pepper [J]. Biotechnology Bulletin, 2024, 40(9): 198-211. |
| [7] | SONG Qian-na, DUAN Yong-hong, FENG Rui-yun. Establishment of CRISPR/Cas9-mediated Highly Efficient Gene Editing System in Microtubers of Potatoes [J]. Biotechnology Bulletin, 2024, 40(9): 33-41. |
| [8] | WANG Ke-ran, YAN Jun-jie, LIU Jian-feng, GAO Yu-lin. Application and Risk of RNAi Technology in Potato Insect Pest Management [J]. Biotechnology Bulletin, 2024, 40(9): 4-10. |
| [9] | ZHANG Xiao-mei, ZHOU Nan-ling, ZHANG Sai-hang, WANG Chao, SHEN Yu-long, GUAN Jun-mei, MA Ling. Cloning and Expression Analysis of StDREBs Gene in Solanum tuberosum L. [J]. Biotechnology Bulletin, 2024, 40(9): 42-50. |
| [10] | MAN Quan-cai, MENG Zi-nuo, LI Wei, CAI Xin-ru, SU Run-dong, FU Chang-qing, GAO Shun-juan, CUI Jiang-hui. Identification and Expression Analysis of AQP Gene Family in Potato [J]. Biotechnology Bulletin, 2024, 40(9): 51-63. |
| [11] | WU Juan, WU Xiao-juan, WANG Pei-jie, XIE Rui, NIE Hu-shuai, LI Nan, MA Yan-hong. Screening and Expression Analysis of ERF Gene Related to Anthocyanin Synthesis in Colored Potato [J]. Biotechnology Bulletin, 2024, 40(9): 82-91. |
| [12] | QIAO Yan, YANG Fang, REN Pan-rong, QI Wei-liang, AN Pei-pei, LI Qian, LI Dan, XIAO Jun-fei. Cloning and Function Analysis of the ScDHNS Gene of Crotonase/Enoyl-CoA Superfamily from a Wild Potato Species [J]. Biotechnology Bulletin, 2024, 40(9): 92-103. |
| [13] | CUI Yuan-yuan, WANG Zhao-yi, BAI Shuang-yu, REN Yu-zhao, DOU Fei-fei, LIU Cai-xia, LIU Feng-lou, WANG Zhang-jun, LI Qing-feng. Genome-wide Identification of Non-specific Phospholipase C Gene Family in Hordeum vulgare L. and Stress Expression Analysis at Seedling Stage [J]. Biotechnology Bulletin, 2024, 40(8): 74-82. |
| [14] | YANG Wei, ZHAO Li-fen, TANG Bing, ZHOU Lin-bi, YANG Juan, MO Chuan-yuan, ZHANG Bao-hui, LI Fei, RUAN Song-lin, DENG Ying. Genome-wide Identification and Expression Analysis of the SRO Gene Family in Brassica juncea L. [J]. Biotechnology Bulletin, 2024, 40(8): 129-141. |
| [15] | ZHOU Ran, WANG Xing-ping, LI Yan-xia, LUORENG Zhuo-ma. Analysis of LncRNA Differential Expression in Mammary Tissue of Cows with Staphylococcus aureus Mastitis [J]. Biotechnology Bulletin, 2024, 40(8): 320-328. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||