








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (9): 51-63.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0439
Previous Articles Next Articles
MAN Quan-cai( ), MENG Zi-nuo, LI Wei, CAI Xin-ru, SU Run-dong, FU Chang-qing, GAO Shun-juan, CUI Jiang-hui(
), MENG Zi-nuo, LI Wei, CAI Xin-ru, SU Run-dong, FU Chang-qing, GAO Shun-juan, CUI Jiang-hui( )
)
Received:2024-05-10
Online:2024-09-26
Published:2024-10-12
Contact:
CUI Jiang-hui
E-mail:15715377671@163.com;cjianghui521@126.com
MAN Quan-cai, MENG Zi-nuo, LI Wei, CAI Xin-ru, SU Run-dong, FU Chang-qing, GAO Shun-juan, CUI Jiang-hui. Identification and Expression Analysis of AQP Gene Family in Potato[J]. Biotechnology Bulletin, 2024, 40(9): 51-63.
| 基因 Gene | 基因ID Gene ID | 染色体位置Chromosome localization | 氨基酸长度Amino acid length/aa | 相对分子量Molecular weight/Da | 理论等电点Theoretical isoelectric point | 亚细胞定位Subcellular localization |
|---|---|---|---|---|---|---|
| StAQP1 | PGSC0003DMG400034503 | Chr01 | 287 | 30698.7 | 6.50 | 质膜Plasma membrane |
| StAQP2 | PGSC0003DMG400000045 | Chr01 | 285 | 30573.2 | 8.84 | 质膜Plasma membrane |
| StAQP3 | PGSC0003DMG400024879 | Chr01 | 112 | 12379.6 | 9.83 | 细胞质Cytoplasm |
| StAQP4 | PGSC0003DMG400001640 | Chr01 | 248 | 26735.5 | 8.21 | 质膜Plasma membrane |
| StAQP5 | PGSC0003DMG400006183 | Chr01 | 283 | 30119.8 | 8.60 | 质膜Plasma membrane |
| StAQP6 | PGSC0003DMG400028456 | Chr02 | 181 | 19461.6 | 6.34 | 液泡Vacuole |
| StAQP7 | PGSC0003DMG401028457 | Chr02 | 290 | 30571 | 9.10 | 质膜Plasma membrane |
| StAQP8 | PGSC0003DMG400003587 | Chr02 | 251 | 26556.8 | 8.08 | 质膜Plasma membrane |
| StAQP9 | PGSC0003DMG400030648 | Chr02 | 267 | 28189.8 | 8.45 | 细胞骨架Cytoskeleton |
| StAQP10 | PGSC0003DMG400027819 | Chr03 | 277 | 29657.2 | 9.55 | 质膜Plasma membrane |
| StAQP11 | PGSC0003DMG400020742 | Chr03 | 286 | 30688.4 | 8.83 | 质膜Plasma membrane |
| StAQP12 | PGSC0003DMG400014818 | Chr03 | 251 | 25951 | 8.67 | 叶绿体Chloroplast |
| StAQP13 | PGSC0003DMG400015275 | Chr03 | 260 | 27777.9 | 9.22 | 细胞质Cytoplasm |
| StAQP14 | PGSC0003DMG400014123 | Chr03 | 306 | 31684.7 | 8.34 | 质膜Plasma membrane |
| StAQP15 | PGSC0003DMG400002552 | Chr03 | 249 | 25161.1 | 6.33 | 细胞质Cytoplasm |
| StAQP16 | PGSC0003DMG401030499 | Chr05 | 272 | 29260.3 | 8.20 | 细胞质Cytoplasm |
| StAQP17 | PGSC0003DMG400023466 | Chr05 | 275 | 29328.2 | 9.65 | 质膜Plasma membrane |
| StAQP18 | PGSC0003DMG400024197 | Chr06 | 286 | 30662.3 | 7.18 | 质膜Plasma membrane |
| StAQP19 | PGSC0003DMG400011875 | Chr06 | 250 | 25151 | 5.48 | 液泡Vacuole |
| StAQP20 | PGSC0003DMG400016528 | Chr06 | 249 | 25089 | 6.06 | 质膜Plasma membrane |
| StAQP21 | PGSC0003DMG400026969 | Chr06 | 259 | 27476.5 | 7.32 | 细胞质Cytoplasm |
| StAQP22 | PGSC0003DMG401005949 | Chr06 | 347 | 37712.9 | 8.32 | 质膜Plasma membrane |
| StAQP23 | PGSC0003DMG400007134 | Chr06 | 250 | 25749.5 | 6.13 | 细胞质Cytoplasm |
| StAQP24 | PGSC0003DMG400030308 | Chr06 | 253 | 26056.9 | 5.48 | 液泡Vacuole |
| StAQP25 | PGSC0003DMG400005780 | Chr08 | 287 | 30790.4 | 7.96 | 质膜Plasma membrane |
| StAQP26 | PGSC0003DMG400005803 | Chr08 | 296 | 30651.2 | 8.48 | 质膜Plasma membrane |
| StAQP27 | PGSC0003DMG400009604 | Chr08 | 247 | 26042.5 | 6.22 | 液泡Vacuole |
| StAQP28 | PGSC0003DMG400012337 | Chr08 | 286 | 30686.6 | 9.34 | 质膜Plasma membrane |
| StAQP29 | PGSC0003DMG402020908 | Chr09 | 196 | 20811.2 | 9.74 | 细胞质Cytoplasm |
| StAQP30 | PGSC0003DMG401020908 | Chr09 | 307 | 32802.8 | 8.11 | 质膜Plasma membrane |
| StAQP31 | PGSC0003DMG400020906 | Chr09 | 279 | 29848.6 | 8.54 | 质膜Plasma membrane |
| StAQP32 | PGSC0003DMG400034609 | Chr10 | 183 | 19587.9 | 8.91 | 液泡Vacuole |
| StAQP33 | PGSC0003DMG400026706 | Chr10 | 262 | 27681.7 | 7.92 | 液泡Vacuole |
| StAQP34 | PGSC0003DMG400026705 | Chr10 | 328 | 35105.8 | 7.47 | 质膜Plasma membrane |
| StAQP35 | PGSC0003DMG400004438 | Chr10 | 326 | 34807.5 | 7.80 | 质膜Plasma membrane |
| StAQP36 | PGSC0003DMG400026718 | Chr10 | 283 | 30409.3 | 9.23 | 质膜Plasma membrane |
| StAQP37 | PGSC0003DMG400010475 | Chr10 | 281 | 30179.7 | 8.42 | 质膜Plasma membrane |
| StAQP38 | PGSC0003DMG400028182 | Chr10 | 248 | 25341.3 | 6.34 | 细胞质Cytoplasm |
| StAQP39 | PGSC0003DMG400008358 | Chr10 | 243 | 25717.4 | 10.24 | 液泡Vacuole |
| StAQP40 | PGSC0003DMG400008078 | Chr11 | 287 | 30628.4 | 7.99 | 质膜Plasma membrane |
| StAQP41 | PGSC0003DMG400018823 | Chr12 | 239 | 25734.9 | 6.67 | 液泡Vacuole |
| StAQP42 | PGSC0003DMG400026463 | Chr12 | 248 | 24998.9 | 5.95 | 液泡Vacuole |
| StAQP43 | PGSC0003DMG400004980 | Chr12 | 288 | 30942.6 | 8.30 | 质膜Plasma membrane |
| StAQP44 | PGSC0003DMG400008556 | Chr12 | 260 | 27844.6 | 8.68 | 液泡Vacuole |
Table 1 Analysis of potato AQP gene family members and physicochemical properties
| 基因 Gene | 基因ID Gene ID | 染色体位置Chromosome localization | 氨基酸长度Amino acid length/aa | 相对分子量Molecular weight/Da | 理论等电点Theoretical isoelectric point | 亚细胞定位Subcellular localization |
|---|---|---|---|---|---|---|
| StAQP1 | PGSC0003DMG400034503 | Chr01 | 287 | 30698.7 | 6.50 | 质膜Plasma membrane |
| StAQP2 | PGSC0003DMG400000045 | Chr01 | 285 | 30573.2 | 8.84 | 质膜Plasma membrane |
| StAQP3 | PGSC0003DMG400024879 | Chr01 | 112 | 12379.6 | 9.83 | 细胞质Cytoplasm |
| StAQP4 | PGSC0003DMG400001640 | Chr01 | 248 | 26735.5 | 8.21 | 质膜Plasma membrane |
| StAQP5 | PGSC0003DMG400006183 | Chr01 | 283 | 30119.8 | 8.60 | 质膜Plasma membrane |
| StAQP6 | PGSC0003DMG400028456 | Chr02 | 181 | 19461.6 | 6.34 | 液泡Vacuole |
| StAQP7 | PGSC0003DMG401028457 | Chr02 | 290 | 30571 | 9.10 | 质膜Plasma membrane |
| StAQP8 | PGSC0003DMG400003587 | Chr02 | 251 | 26556.8 | 8.08 | 质膜Plasma membrane |
| StAQP9 | PGSC0003DMG400030648 | Chr02 | 267 | 28189.8 | 8.45 | 细胞骨架Cytoskeleton |
| StAQP10 | PGSC0003DMG400027819 | Chr03 | 277 | 29657.2 | 9.55 | 质膜Plasma membrane |
| StAQP11 | PGSC0003DMG400020742 | Chr03 | 286 | 30688.4 | 8.83 | 质膜Plasma membrane |
| StAQP12 | PGSC0003DMG400014818 | Chr03 | 251 | 25951 | 8.67 | 叶绿体Chloroplast |
| StAQP13 | PGSC0003DMG400015275 | Chr03 | 260 | 27777.9 | 9.22 | 细胞质Cytoplasm |
| StAQP14 | PGSC0003DMG400014123 | Chr03 | 306 | 31684.7 | 8.34 | 质膜Plasma membrane |
| StAQP15 | PGSC0003DMG400002552 | Chr03 | 249 | 25161.1 | 6.33 | 细胞质Cytoplasm |
| StAQP16 | PGSC0003DMG401030499 | Chr05 | 272 | 29260.3 | 8.20 | 细胞质Cytoplasm |
| StAQP17 | PGSC0003DMG400023466 | Chr05 | 275 | 29328.2 | 9.65 | 质膜Plasma membrane |
| StAQP18 | PGSC0003DMG400024197 | Chr06 | 286 | 30662.3 | 7.18 | 质膜Plasma membrane |
| StAQP19 | PGSC0003DMG400011875 | Chr06 | 250 | 25151 | 5.48 | 液泡Vacuole |
| StAQP20 | PGSC0003DMG400016528 | Chr06 | 249 | 25089 | 6.06 | 质膜Plasma membrane |
| StAQP21 | PGSC0003DMG400026969 | Chr06 | 259 | 27476.5 | 7.32 | 细胞质Cytoplasm |
| StAQP22 | PGSC0003DMG401005949 | Chr06 | 347 | 37712.9 | 8.32 | 质膜Plasma membrane |
| StAQP23 | PGSC0003DMG400007134 | Chr06 | 250 | 25749.5 | 6.13 | 细胞质Cytoplasm |
| StAQP24 | PGSC0003DMG400030308 | Chr06 | 253 | 26056.9 | 5.48 | 液泡Vacuole |
| StAQP25 | PGSC0003DMG400005780 | Chr08 | 287 | 30790.4 | 7.96 | 质膜Plasma membrane |
| StAQP26 | PGSC0003DMG400005803 | Chr08 | 296 | 30651.2 | 8.48 | 质膜Plasma membrane |
| StAQP27 | PGSC0003DMG400009604 | Chr08 | 247 | 26042.5 | 6.22 | 液泡Vacuole |
| StAQP28 | PGSC0003DMG400012337 | Chr08 | 286 | 30686.6 | 9.34 | 质膜Plasma membrane |
| StAQP29 | PGSC0003DMG402020908 | Chr09 | 196 | 20811.2 | 9.74 | 细胞质Cytoplasm |
| StAQP30 | PGSC0003DMG401020908 | Chr09 | 307 | 32802.8 | 8.11 | 质膜Plasma membrane |
| StAQP31 | PGSC0003DMG400020906 | Chr09 | 279 | 29848.6 | 8.54 | 质膜Plasma membrane |
| StAQP32 | PGSC0003DMG400034609 | Chr10 | 183 | 19587.9 | 8.91 | 液泡Vacuole |
| StAQP33 | PGSC0003DMG400026706 | Chr10 | 262 | 27681.7 | 7.92 | 液泡Vacuole |
| StAQP34 | PGSC0003DMG400026705 | Chr10 | 328 | 35105.8 | 7.47 | 质膜Plasma membrane |
| StAQP35 | PGSC0003DMG400004438 | Chr10 | 326 | 34807.5 | 7.80 | 质膜Plasma membrane |
| StAQP36 | PGSC0003DMG400026718 | Chr10 | 283 | 30409.3 | 9.23 | 质膜Plasma membrane |
| StAQP37 | PGSC0003DMG400010475 | Chr10 | 281 | 30179.7 | 8.42 | 质膜Plasma membrane |
| StAQP38 | PGSC0003DMG400028182 | Chr10 | 248 | 25341.3 | 6.34 | 细胞质Cytoplasm |
| StAQP39 | PGSC0003DMG400008358 | Chr10 | 243 | 25717.4 | 10.24 | 液泡Vacuole |
| StAQP40 | PGSC0003DMG400008078 | Chr11 | 287 | 30628.4 | 7.99 | 质膜Plasma membrane |
| StAQP41 | PGSC0003DMG400018823 | Chr12 | 239 | 25734.9 | 6.67 | 液泡Vacuole |
| StAQP42 | PGSC0003DMG400026463 | Chr12 | 248 | 24998.9 | 5.95 | 液泡Vacuole |
| StAQP43 | PGSC0003DMG400004980 | Chr12 | 288 | 30942.6 | 8.30 | 质膜Plasma membrane |
| StAQP44 | PGSC0003DMG400008556 | Chr12 | 260 | 27844.6 | 8.68 | 液泡Vacuole |
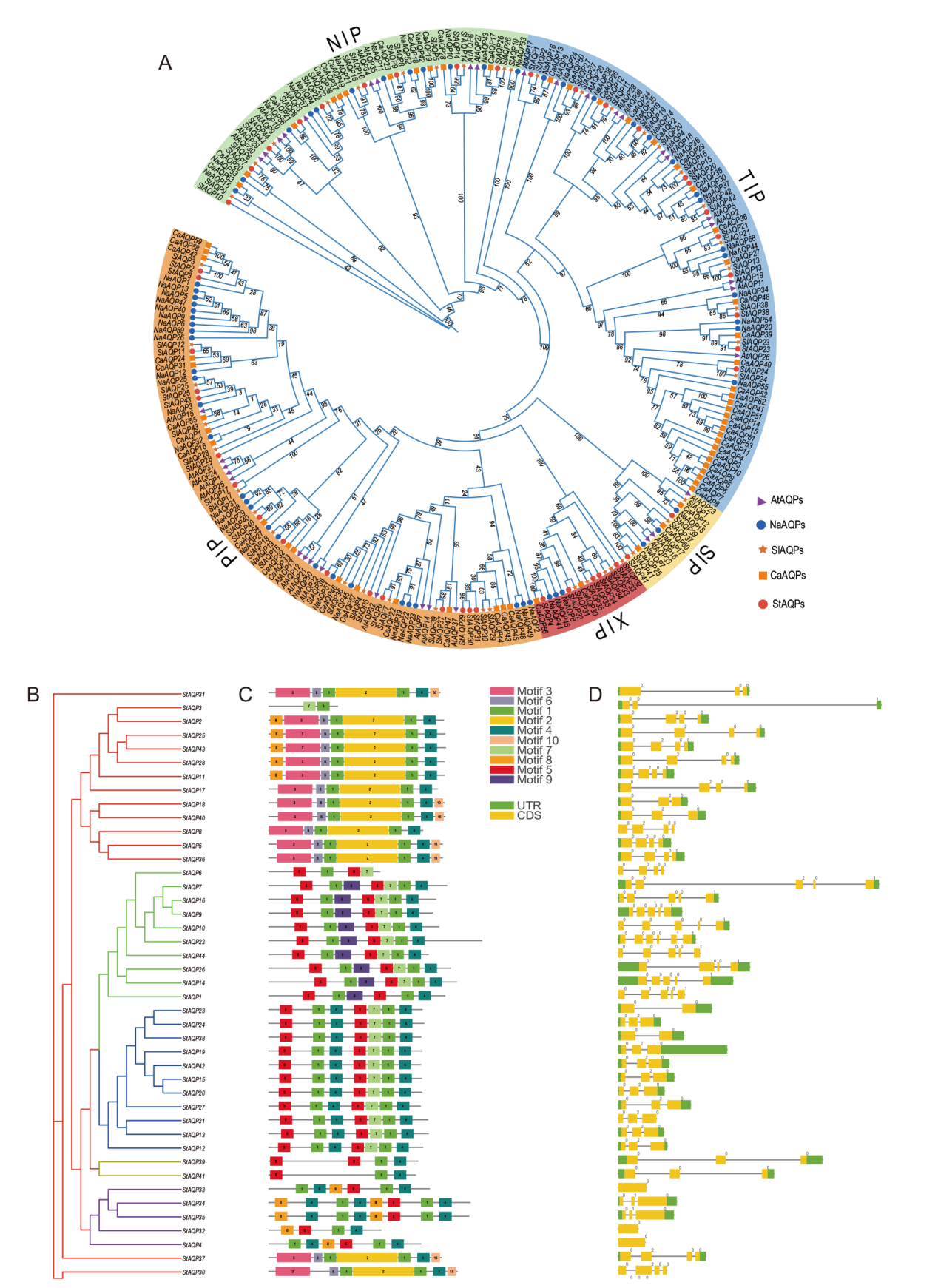
Fig. 1 Phylogenetic, gene structure and conserved motif analysis of the AQP gene family A: Phylogenetic trees of AQP families in the A. thaliana, N. attenuata, S. tuberosum, S. lycopersicum, and Capsicum annuum. B: Phylogenetic tree of the StAQP gene family. C: Conserved motif analysis of the StAQP gene. D: Analysis of StAQP gene structure

Fig. 3 Gene duplication events in the AQP family of S. tuberosum A: Chromosomal localization map of StAQP gene. B: Collinearity analysis of StAQP gene. C: Collinearity analysis of AQP genes in multiple species(At: A. thaliana; St: S. tuberosum; Na: N. attenuata; Ca: C. annuum; Sl: S. lycopersicum)
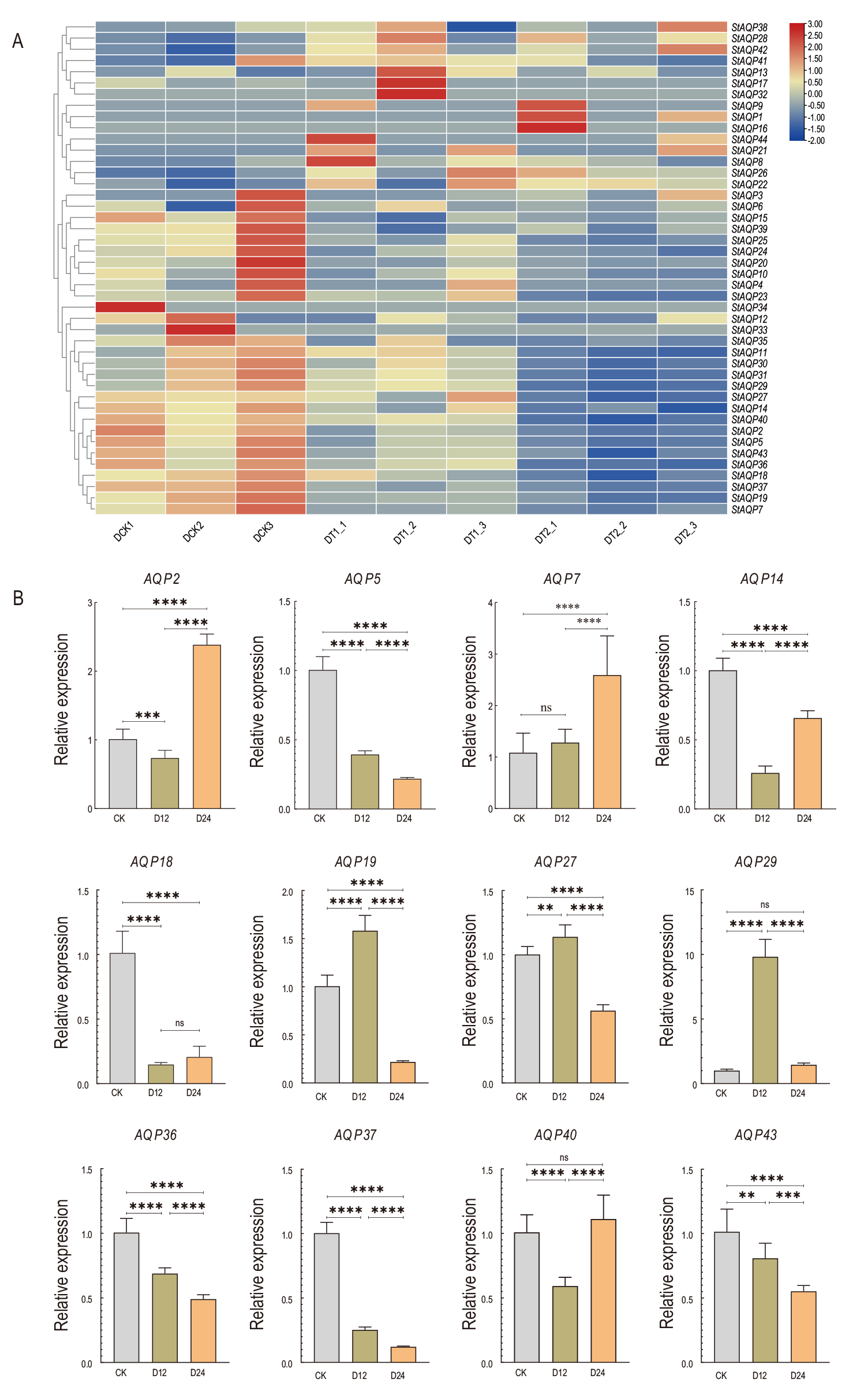
Fig. 5 Expression of StAQP gene under drought stress A: Expression of StAQP under drought stress. B: Relative expression of StAQP under drought stress, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. The same below
| [1] | Kopecká R, Kameniarová M, Černý M, et al. Abiotic stress in crop production[J]. Int J Mol Sci, 2023, 24(7): 6603. |
| [2] |
Chen XX, Ding YL, Yang YQ, et al. Protein kinases in plant responses to drought, salt, and cold stress[J]. J Integr Plant Biol, 2021, 63(1): 53-78.
doi: 10.1111/jipb.13061 |
| [3] | Dietz KJ, Zörb C, Geilfus CM. Drought and crop yield[J]. Plant Biol, 2021, 23(6): 881-893. |
| [4] | Riyazuddin R, Nisha NS, Singh K, et al. Involvement of dehydrin proteins in mitigating the negative effects of drought stress in plants[J]. Plant Cell Rep, 2022, 41(3): 519-533. |
| [5] | Sánchez-Bermúdez M, Del Pozo JC, Pernas M. Effects of combined abiotic stresses related to climate change on root growth in crops[J]. Front Plant Sci, 2022, 13: 918537. |
| [6] | Zhao SS, Zhang QK, Liu MY, et al. Regulation of plant responses to salt stress[J]. Int J Mol Sci, 2021, 22(9): 4609. |
| [7] | Kausar R, Komatsu S. Proteomic approaches to uncover salt stress response mechanisms in crops[J]. Int J Mol Sci, 2022, 24(1): 518. |
| [8] | Colin L, Ruhnow F, Zhu JK, et al. The cell biology of primary cell walls during salt stress[J]. Plant Cell, 2023, 35(1): 201-217. |
| [9] | Xu J, Li Y, Kaur L, et al. Functional food based on potato[J]. Foods, 2023, 12(11): 2145. |
| [10] | Hill D, Nelson D, Hammond J, et al. Morphophysiology of potato(Solanum tuberosum)in response to drought stress: paving the way forward[J]. Front Plant Sci, 2021, 11: 597554. |
| [11] | Li Q, Qin YZ, Hu XX, et al. Physiology and gene expression analysis of potato(Solanum tuberosum L.) in salt stress[J]. Plants, 2022, 11(12): 1565. |
| [12] | Wang Y, Zhao ZJ, Liu F, et al. Versatile roles of aquaporins in plant growth and development[J]. Int J Mol Sci, 2020, 21(24): 9485. |
| [13] |
Li SC, Li CL, Wang WD. Molecular aspects of aquaporins[J]. Vitam Horm, 2020, 113: 129-181.
doi: S0083-6729(19)30079-2 pmid: 32138947 |
| [14] | Vats S, Sudhakaran S, Bhardwaj A, et al. Targeting aquaporins to alleviate hazardous metal(loid)s imposed stress in plants[J]. J Hazard Mater, 2021, 408: 124910. |
| [15] | Byrt CS, Zhang RY, Magrath I, et al. Exploring aquaporin functions during changes in leaf water potential[J]. Front Plant Sci, 2023, 14: 1213454. |
| [16] |
Patel J, Mishra A. Plant aquaporins alleviate drought tolerance in plants by modulating cellular biochemistry, root-architecture, and photosynthesis[J]. Physiol Plant, 2021, 172(2): 1030-1044.
doi: 10.1111/ppl.13324 pmid: 33421148 |
| [17] |
Ahmed S, Kouser S, Asgher M, et al. Plant aquaporins: a frontward to make crop plants drought resistant[J]. Physiol Plant, 2021, 172(2): 1089-1105.
doi: 10.1111/ppl.13416 pmid: 33826759 |
| [18] | Kumari A, Bhatla SC. Regulation of salt-stressed sunflower(Helianthus annuus)seedling's water status by the coordinated action of Na+/K+ accumulation, nitric oxide, and aquaporin expression[J]. Funct Plant Biol, 2021, 48(6): 573-587. |
| [19] | Feng ZJ, Xu SC, Liu N, et al. Identification of the AQP members involved in abiotic stress responses from Arabidopsis[J]. Gene, 2018, 646: 64-73. |
| [20] | De Rosa A, Watson-Lazowski A, Evans JR, et al. Genome-wide identification and characterisation of aquaporins in Nicotiana tabacum and their relationships with other Solanaceae species[J]. BMC Plant Biol, 2020, 20(1): 266. |
| [21] | Yi XF, Sun XC, Tian R, et al. Genome-wide characterization of the aquaporin gene family in radish and functional analysis of RsPIP2-6 involved in salt stress[J]. Front Plant Sci, 2022, 13: 860742. |
| [22] |
Stajich JE. An introduction to BioPerl[J]. Methods Mol Biol, 2007, 406: 535-548.
pmid: 18287711 |
| [23] | Horton P, Park KJ, Obayashi T, et al. WoLF PSORT: protein localization predictor[J]. Nucleic Acids Res, 2007, 35(Web Server issue): W585-W587. |
| [24] | Nguyen LT, Schmidt HA, von Haeseler A, et al. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies[J]. Mol Biol Evol, 2015, 32(1): 268-274. |
| [25] | Letunic I, Bork P. Interactive Tree Of Life(iTOL)v5: an online tool for phylogenetic tree display and annotation[J]. Nucleic Acids Res, 2021, 49(W1): W293-W296. |
| [26] |
Hu B, Jin JP, Guo AY, et al. GSDS 2.0: an upgraded gene feature visualization server[J]. Bioinformatics, 2015, 31(8): 1296-1297.
doi: 10.1093/bioinformatics/btu817 pmid: 25504850 |
| [27] | Bailey TL, Boden M, Buske FA, et al. MEME SUITE: tools for motif discovery and searching[J]. Nucleic Acids Res, 2009, 37(Web Server issue): W202-W208. |
| [28] |
Lescot M, Déhais P, Thijs G, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences[J]. Nucleic Acids Res, 2002, 30(1): 325-327.
doi: 10.1093/nar/30.1.325 pmid: 11752327 |
| [29] | Wang YP, Tang HB, Debarry JD, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity[J]. Nucleic Acids Res, 2012, 40(7): e49. |
| [30] |
Krzywinski M, Schein J, Birol I, et al. Circos: an information aesthetic for comparative genomics[J]. Genome Res, 2009, 19(9): 1639-1645.
doi: 10.1101/gr.092759.109 pmid: 19541911 |
| [31] | Wang DP, Zhang YB, Zhang Z, et al. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies[J]. Genomics Proteomics Bioinformatics, 2010, 8(1): 77-80. |
| [32] | Szklarczyk D, Kirsch R, Koutrouli M, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest[J]. Nucleic Acids Res, 2023, 51(D1): D638-D646. |
| [33] | Franz M, Lopes CT, Fong D, et al. Cytoscape.js 2023 update: a graph theory library for visualization and analysis[J]. Bioinformatics, 2023, 39(1): btad031. |
| [34] | Chen CJ, Wu Y, Li JW, et al. TBtools-II: a “one for all, all for one” bioinformatics platform for biological big-data mining[J]. Mol Plant, 2023, 16(11): 1733-1742. |
| [35] |
Tang X, Zhang N, Si HJ, et al. Selection and validation of reference genes for RT-qPCR analysis in potato under abiotic stress[J]. Plant Methods, 2017, 13: 85.
doi: 10.1186/s13007-017-0238-7 pmid: 29075311 |
| [36] |
Maren NA, Duduit JR, Huang DB, et al. Stepwise optimization of real-time RT-PCR analysis[J]. Methods Mol Biol, 2023, 2653: 317-332.
doi: 10.1007/978-1-0716-3131-7_20 pmid: 36995635 |
| [37] | Ishibashi K, Kondo S, Hara S, et al. The evolutionary aspects of aquaporin family[J]. Am J Physiol Regul Integr Comp Physiol, 2011, 300(3): R566-R576. |
| [38] |
Maurel C, Boursiac Y, Luu DT, et al. Aquaporins in plants[J]. Physiol Rev, 2015, 95(4): 1321-1358.
doi: 10.1152/physrev.00008.2015 pmid: 26336033 |
| [39] | Yaghobi M, Heidari P. Genome-wide analysis of aquaporin gene family in Triticum turgidum and its expression profile in response to salt stress[J]. Genes, 2023, 14(1): 202. |
| [40] | He W, Liu MY, Qin XY, et al. Genome-wide identification and expression analysis of the aquaporin gene family in Lycium barbarum during fruit ripening and seedling response to heat stress[J]. Curr Issues Mol Biol, 2022, 44(12): 5933-5948. |
| [41] |
Wang R, Li RZ, Cheng LN, et al. SlERF52 regulates SlTIP1;1 expression to accelerate tomato pedicel abscission[J]. Plant Physiol, 2021, 185(4): 1829-1846.
doi: 10.1093/plphys/kiab026 pmid: 33638643 |
| [1] | SHEN Peng, GAO Ya-Bin, DING Hong. Identification and Expression Analysis of SAT Gene Family in Potato(Solanum tuberosum L.) [J]. Biotechnology Bulletin, 2024, 40(9): 64-73. |
| [2] | WANG Chao, BAI Ru-qian, GUAN Jun-mei, LUO Ji-lin, HE Xue-jiao, CHI Shao-yi, MA Ling. Promotion of StHY5 in the Synthesis of SGAs during Tuber Turning-green of Potato [J]. Biotechnology Bulletin, 2024, 40(9): 113-122. |
| [3] | XIA Shi-xuan, GENG Ze-dong, ZHU Guang-tao, ZHANG Chun-zhi, LI Da-wei. Quick Detection of Potato Pollen Viability Based on Deep Learning [J]. Biotechnology Bulletin, 2024, 40(9): 123-130. |
| [4] | MAO Xiang-hong, LU Yao, FAN Xiang-bin, DU Pei-bing, BAI Xiao-dong. Genetic Diversity Analysis of Potato Varieties Based on SSR Fluorescent Marker Capillary Electrophoresis and Construction of Molecular Identity Card [J]. Biotechnology Bulletin, 2024, 40(9): 131-140. |
| [5] | YUAN Lan, HUANG Ya-nan, ZHANG Bei-ni, XIONG Yu-meng, WANG Hong-yang. High-throughput Sample Preparation Method for the Identification of Potato Ploidy Using Flow Cytometry [J]. Biotechnology Bulletin, 2024, 40(9): 141-147. |
| [6] | WU Hui-qin, WANG Yan-hong, LIU Han, SI Zheng, LIU Xue-qing, WANG Jing, YANG Yi, CHENG Yan. Identification and Expression Analysis of UGT Gene Family in Pepper [J]. Biotechnology Bulletin, 2024, 40(9): 198-211. |
| [7] | TAN Bo-wen, ZHANG Yi, ZHANG Peng, WANG Zhen-yu, MA Qiu-xiang. Identification and Bioinformatics Analysis of Gene in the Magnesium Transporter Family in Cassava [J]. Biotechnology Bulletin, 2024, 40(9): 20-32. |
| [8] | SONG Qian-na, DUAN Yong-hong, FENG Rui-yun. Establishment of CRISPR/Cas9-mediated Highly Efficient Gene Editing System in Microtubers of Potatoes [J]. Biotechnology Bulletin, 2024, 40(9): 33-41. |
| [9] | WANG Ke-ran, YAN Jun-jie, LIU Jian-feng, GAO Yu-lin. Application and Risk of RNAi Technology in Potato Insect Pest Management [J]. Biotechnology Bulletin, 2024, 40(9): 4-10. |
| [10] | ZHANG Xiao-mei, ZHOU Nan-ling, ZHANG Sai-hang, WANG Chao, SHEN Yu-long, GUAN Jun-mei, MA Ling. Cloning and Expression Analysis of StDREBs Gene in Solanum tuberosum L. [J]. Biotechnology Bulletin, 2024, 40(9): 42-50. |
| [11] | WU Juan, WU Xiao-juan, WANG Pei-jie, XIE Rui, NIE Hu-shuai, LI Nan, MA Yan-hong. Screening and Expression Analysis of ERF Gene Related to Anthocyanin Synthesis in Colored Potato [J]. Biotechnology Bulletin, 2024, 40(9): 82-91. |
| [12] | QIAO Yan, YANG Fang, REN Pan-rong, QI Wei-liang, AN Pei-pei, LI Qian, LI Dan, XIAO Jun-fei. Cloning and Function Analysis of the ScDHNS Gene of Crotonase/Enoyl-CoA Superfamily from a Wild Potato Species [J]. Biotechnology Bulletin, 2024, 40(9): 92-103. |
| [13] | SONG Bing-fang, LIU Ning, CHENG Xin-yan, XU Xiao-bin, TIAN Wen-mao, GAO Yue, BI Yang, WANG Yi. Identification of Potato G6PDH Gene Family and Its Expression Analysis in Damaged Tubers [J]. Biotechnology Bulletin, 2024, 40(9): 104-112. |
| [14] | ZHOU Ran, WANG Xing-ping, LI Yan-xia, LUORENG Zhuo-ma. Analysis of LncRNA Differential Expression in Mammary Tissue of Cows with Staphylococcus aureus Mastitis [J]. Biotechnology Bulletin, 2024, 40(8): 320-328. |
| [15] | WU Shuai, XIN Yan-ni, MAI Chun-hai, MU Xiao-ya, WANG Min, YUE Ai-qin, ZHAO Jin-zhong, WU Shen-jie, DU Wei-jun, WANG Li-xiang. Genome-wide Identification and Stress Response Analysis of Soybean GS Gene Family [J]. Biotechnology Bulletin, 2024, 40(8): 63-73. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||