








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (1): 157-172.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0656
Previous Articles Next Articles
HE Cai-lin1,2( ), LU Jing1,2, GUO Hui-hui1,2, LI Xiao-an1,2, WU Qi1,2(
), LU Jing1,2, GUO Hui-hui1,2, LI Xiao-an1,2, WU Qi1,2( )
)
Received:2024-07-10
Online:2025-01-26
Published:2025-01-22
Contact:
WU Qi
E-mail:2737141093@qq.com;jerviswuqi@126.com
HE Cai-lin, LU Jing, GUO Hui-hui, LI Xiao-an, WU Qi. Genome-wide Identification and Expression Analysis of the MADS-box Gene Family in Quinoa[J]. Biotechnology Bulletin, 2025, 41(1): 157-172.
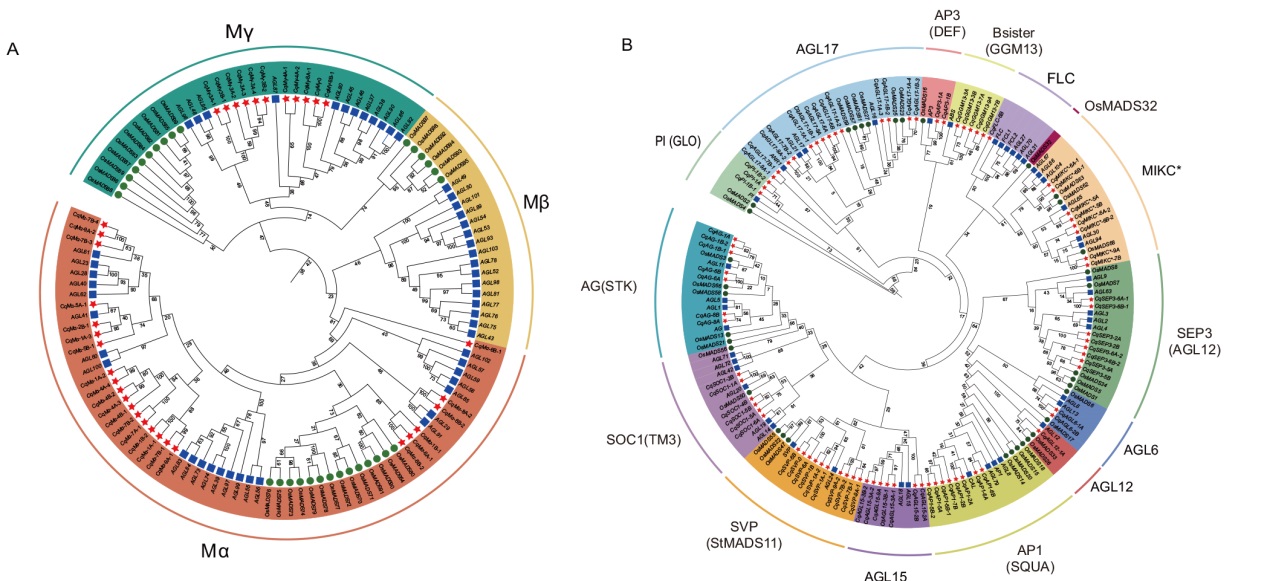
Fig. 1 MADS-box protein phylogenetic tree of quinoa, rice and Arabidopsis A: Phylogenetic tree of type I MADS-box proteins in quinoa(Chenopodium quinoa), Arabidopsis and rice(Oryza sativa). B: Phylogenetic tree of type II MADS-box proteins in quinoa, Arabidopsis and rice. The red five-pointed star indicates quinoa protein, the purple box indicates Arabidopsis protein, and the green circle indicates rice protein
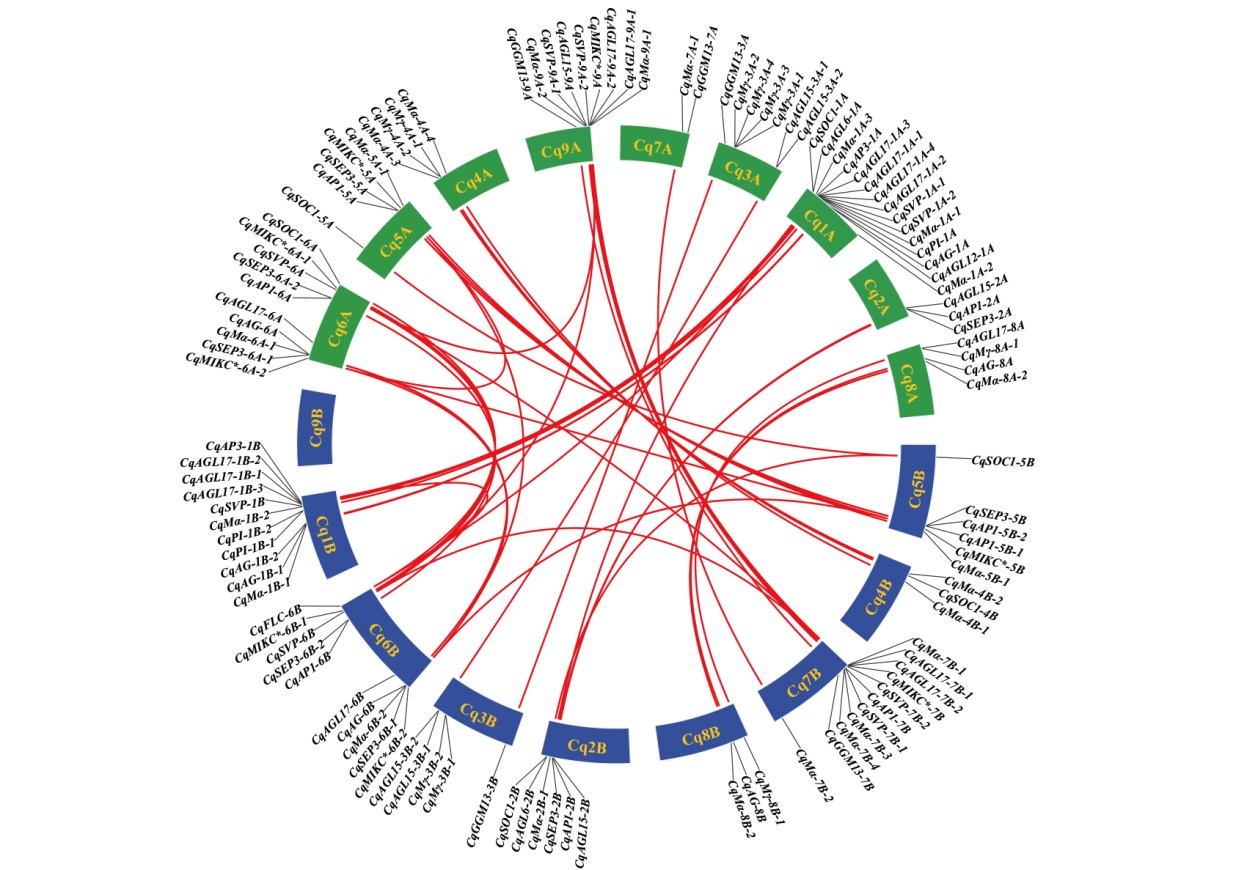
Fig. 4 Collinearity between the MADS-box of quinoa sub-genome A and sub-genome B Green indicate quinoa A sub-genome, and blue represents quinoa B sub-genome

Fig. 5 Collinearity analysis of quinoa MADS-box members with that in C. pallidicaule(A)and C. suecicum(B) Green indicates quinoa chromosome, purple indicates C. pallidicaule chromosome and blue indicates C. suecicum chromosome
| 基因名称 Gene name | 基因 ID Gene ID | 染色体 Chromosome | 起始位置 Start-end position(+/-strand) | 氨基酸数 Number of amino acids/aa | 类型 Type |
|---|---|---|---|---|---|
| CqSVP-1A-2 | CQ048095 | Cq1A | 1 174 6420-11 746 605(-) | 61 | 假基因-串联复制 |
| CqAGL17-1A-1 | CQ047997 | Cq1A | 10 659 725-10 659 946(-) | 73 | 假基因-串联复制 |
| CqAGL12-1A | CQ048800 | Cq1A | 22 590 418-22 590 639(-) | 73 | 假基因-节段复制 |
| CqAGL17-1A-2 | CQ048000 | Cq1A | 10 725 903-10 726 115(-) | 70 | 假基因-串联复制 |
| CqAG-1B-2 | CQ023885 | Cq1B | 49 507 013-49 507 303(-) | 96 | 假基因-串联复制 |
| CqMα-1B-1 | CQ023867 | Cq1B | 48 895 860-48 896 081(-) | 73 | 假基因-节段复制 |
| CqAGL17-1B-1 | CQ024731 | Cq1B | 64 342 030-64 342 293(+) | 87 | 假基因-串联复制 |
| CqAGL15-3B-2 | CQ019107 | Cq3B | 66 834 322-66 834 510(-) | 62 | 假基因-串联复制 |
| CqAGL15-3B-1 | CQ019105 | Cq3B | 66 809 225-66 812 527(-) | 99 | 假基因-节段复制 |
| CqSEP3-5B | CQ005895 | Cq5B | 67 053 435-67 053 671(+) | 78 | 假基因-串联复制 |
| CqAP1-5B-2 | CQ005897 | Cq5B | 67 131 352-67 131 611(+) | 64 | 假基因-串联复制 |
| CqMα-6A-1 | CQ025753 | Cq6A | 4 153 499-4 153 970(+) | 74 | 假基因-串联复制 |
| CqMα-6B-2 | CQ000301 | Cq6B | 3 428 220-3 429 042(+) | 75 | 假基因-节段复制 |
| CqSVP-7B-1 | CQ007326 | Cq7B | 3 310 550-3 310 735(-) | 61 | 假基因-串联复制 |
| CqMIKC*-7B | CQ007095 | Cq7B | 1 279 202-1 286 618(-) | 97 | 假基因-串联复制 |
| CqSVP-9A-1 | CQ052433 | Cq9A | 51 460 017-51 462 248(+) | 73 | 假基因-串联复制 |
| CqAGL17-9A-1 | CQ052648 | Cq9A | 53 340 285-53 346 018(+) | 77 | 假基因-串联复制 |
Table 1 Information of pseudogenes in the MADS-box family of quinoa
| 基因名称 Gene name | 基因 ID Gene ID | 染色体 Chromosome | 起始位置 Start-end position(+/-strand) | 氨基酸数 Number of amino acids/aa | 类型 Type |
|---|---|---|---|---|---|
| CqSVP-1A-2 | CQ048095 | Cq1A | 1 174 6420-11 746 605(-) | 61 | 假基因-串联复制 |
| CqAGL17-1A-1 | CQ047997 | Cq1A | 10 659 725-10 659 946(-) | 73 | 假基因-串联复制 |
| CqAGL12-1A | CQ048800 | Cq1A | 22 590 418-22 590 639(-) | 73 | 假基因-节段复制 |
| CqAGL17-1A-2 | CQ048000 | Cq1A | 10 725 903-10 726 115(-) | 70 | 假基因-串联复制 |
| CqAG-1B-2 | CQ023885 | Cq1B | 49 507 013-49 507 303(-) | 96 | 假基因-串联复制 |
| CqMα-1B-1 | CQ023867 | Cq1B | 48 895 860-48 896 081(-) | 73 | 假基因-节段复制 |
| CqAGL17-1B-1 | CQ024731 | Cq1B | 64 342 030-64 342 293(+) | 87 | 假基因-串联复制 |
| CqAGL15-3B-2 | CQ019107 | Cq3B | 66 834 322-66 834 510(-) | 62 | 假基因-串联复制 |
| CqAGL15-3B-1 | CQ019105 | Cq3B | 66 809 225-66 812 527(-) | 99 | 假基因-节段复制 |
| CqSEP3-5B | CQ005895 | Cq5B | 67 053 435-67 053 671(+) | 78 | 假基因-串联复制 |
| CqAP1-5B-2 | CQ005897 | Cq5B | 67 131 352-67 131 611(+) | 64 | 假基因-串联复制 |
| CqMα-6A-1 | CQ025753 | Cq6A | 4 153 499-4 153 970(+) | 74 | 假基因-串联复制 |
| CqMα-6B-2 | CQ000301 | Cq6B | 3 428 220-3 429 042(+) | 75 | 假基因-节段复制 |
| CqSVP-7B-1 | CQ007326 | Cq7B | 3 310 550-3 310 735(-) | 61 | 假基因-串联复制 |
| CqMIKC*-7B | CQ007095 | Cq7B | 1 279 202-1 286 618(-) | 97 | 假基因-串联复制 |
| CqSVP-9A-1 | CQ052433 | Cq9A | 51 460 017-51 462 248(+) | 73 | 假基因-串联复制 |
| CqAGL17-9A-1 | CQ052648 | Cq9A | 53 340 285-53 346 018(+) | 77 | 假基因-串联复制 |

Fig. 6 Prediction of cis-acting elements in the promoter region of quinoa MADS-box family members A: The distribution map of cis-acting elements in the promoter region of quinoa MADS-box family members. B: Statistics on the number of cis-acting elements in the promoter region of quinoa MADS-box family members

Fig. 7 Expression analysis of MADS-box in different tissues, six-stage inflorescences, and seed germination A: Expressions of MADS-box in different tissues. The expression value is represented by the FPKM values generated from the RNA-seq data(the same below). B: Expression of MADS-box in quinoa inflorescence development. YP1-YP4 indicates unbranched young spikelets in the early developmental stage. P1 and P2 indicate branched panicles at 2 stages of late development. C: Expression changes of MADS-box during seed germination. BL-A-5h/-15h and BL-5h/-15h indicate the ABA-treated and untreated BL materials at 5 h and 15 h after germination, respectively. Each expression value is generated from three replicates. The expressions of the same gene in the 6 stages were normalized to 0 to 1 by row
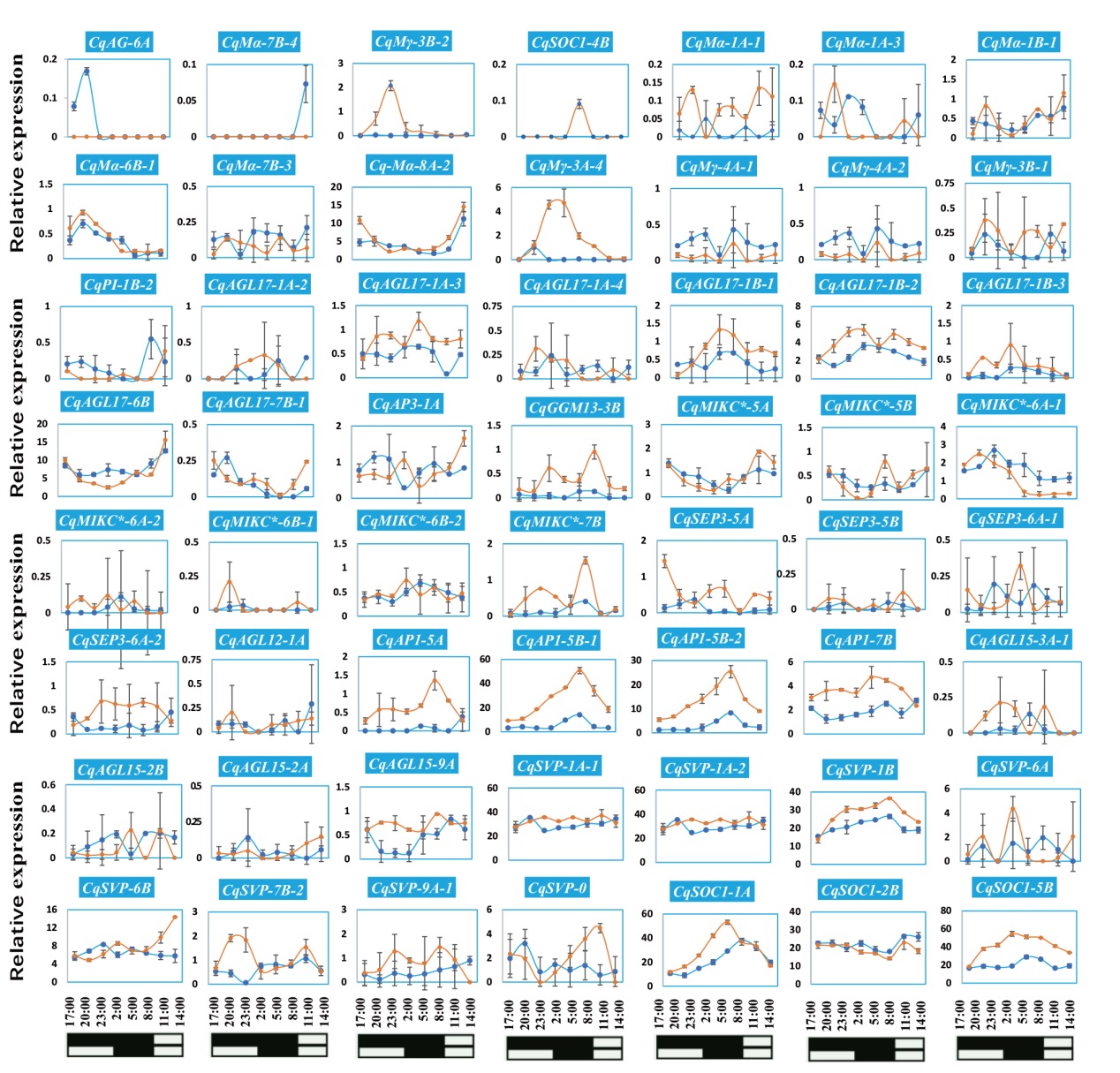
Fig. 8 Diurnal expression patterns of CqMADS-box under short- and long-day conditions Leaf samples of quinoa seedlings grown for two weeks under short-day and long-day conditions were collected at 17:00, 20:00, 23:00, 02:00, 05:00, 08:00, 11:00 and 14:00, respectively. RNA-seq is used to analyze the expression pattern of CqMADS-box. The solid blue line is the gene expression curve under LD, and the solid orange line is the gene expression curve under SD
| [1] |
López-Marqués RL, Nørrevang AF, Ache P, et al. Prospects for the accelerated improvement of the resilient crop quinoa[J]. J Exp Bot, 2020, 71(18): 5333-5347.
doi: 10.1093/jxb/eraa285 pmid: 32643753 |
| [2] | Patiranage DSR, Asare E, Maldonado-Taipe N, et al. Haplotype variations of major flowering time genes in quinoa unveil their role in the adaptation to different environmental conditions[J]. Plant Cell Environ, 2021, 44(8): 2565-2579. |
| [3] |
Benlloch R, Berbel A, Ali L, et al. Genetic control of inflorescence architecture in legumes[J]. Front Plant Sci, 2015, 6: 543.
doi: 10.3389/fpls.2015.00543 pmid: 26257753 |
| [4] |
Schilling S, Pan SR, Kennedy A, et al. MADS-box genes and crop domestication: the jack of all traits[J]. J Exp Bot, 2018, 69(7): 1447-1469.
doi: 10.1093/jxb/erx479 pmid: 29474735 |
| [5] | Alvarez-Buylla ER, Pelaz S, Liljegren SJ, et al. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals[J]. Proc Natl Acad Sci U S A, 2000, 97(10): 5328-5333. |
| [6] |
Kaufmann K, Melzer R, Theissen G. MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants[J]. Gene, 2005, 347(2): 183-198.
doi: 10.1016/j.gene.2004.12.014 pmid: 15777618 |
| [7] |
Riechmann JL, Wang M, Meyerowitz EM. DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS[J]. Nucleic Acids Res, 1996, 24(16): 3134-3141.
pmid: 8774892 |
| [8] | Melzer R, Verelst W, Theiβen G. The class E floral homeotic protein SEPALLATA3 is sufficient to loop DNA in ‘floral quartet’-like complexes in vitro[J]. Nucleic Acids Res, 2009, 37(1): 144-157. |
| [9] | Honma T, Goto K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs[J]. Nature, 2001, 409(6819): 525-529. |
| [10] |
Wang YY, Jiang H, Wang GF. PHERES1 controls endosperm gene imprinting and seed development[J]. Trends Plant Sci, 2020, 25(6): 517-519.
doi: S1360-1385(20)30086-8 pmid: 32407690 |
| [11] | Paul P, Dhatt BK, Miller M, et al. MADS78 and MADS79 are essential regulators of early seed development in rice[J]. Plant Physiol, 2020, 182(2): 933-948. |
| [12] | Zhang JL, Dong TT, Hu ZL, et al. A SEPALLATA MADS-box transcription factor, SlMBP21, functions as a negative regulator of flower number and fruit yields in tomato[J]. Plants, 2024, 13(10): 1421. |
| [13] |
Amasino RM, Michaels SD. The timing of flowering[J]. Plant Physiol, 2010, 154(2): 516-520.
doi: 10.1104/pp.110.161653 pmid: 20921176 |
| [14] | Liu Y, Guan CN, Chen YY, et al. Evolutionary analysis of MADS-box genes in buckwheat species and functional study of FdMADS28 in flavonoid metabolism[J]. Plant Physiol Biochem, 2024, 210: 108637. |
| [15] |
Rey E, Maughan PJ, Maumus F, et al. A chromosome-scale assembly of the quinoa genome provides insights into the structure and dynamics of its subgenomes[J]. Commun Biol, 2023, 6(1): 1263.
doi: 10.1038/s42003-023-05613-4 pmid: 38092895 |
| [16] | Wu Q, Bai X, Wu XY, et al. Transcriptome profiling identifies transcription factors and key homologs involved in seed dormancy and germination regulation of Chenopodium quinoa[J]. Plant Physiol Biochem, 2020, 151: 443-456. |
| [17] | Wu Q, Bai X, Zhao W, et al. Investigation into the underlying regulatory mechanisms shaping inflorescence architecture in Chenopo-dium quinoa[J]. BMC Genomics, 2019, 20(1): 658. |
| [18] | Wu Q, Luo YM, Wu XY, et al. Identification of the specific long-noncoding RNAs involved in night-break mediated flowering retardation in Chenopodium quinoa[J]. BMC Genomics, 2021, 22(1): 284. |
| [19] |
Albert VA, Krabbenhoft TJ. Navigating the CoGe online software suite for polyploidy research[J]. Methods Mol Biol, 2023, 2545: 19-45.
doi: 10.1007/978-1-0716-2561-3_2 pmid: 36720806 |
| [20] | Garcia-Hernandez M, Berardini TZ, Chen GH, et al. TAIR: a resource for integrated Arabidopsis data[J]. Funct Integr Genomics, 2002, 2(6): 239-253. |
| [21] |
Parenicová L, de Folter S, Kieffer M, et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world[J]. Plant Cell, 2003, 15(7): 1538-1551.
doi: 10.1105/tpc.011544 pmid: 12837945 |
| [22] |
Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant, 2020, 13(8): 1194-1202.
doi: S1674-2052(20)30187-8 pmid: 32585190 |
| [23] | Mistry J, Chuguransky S, Williams L, et al. Pfam: the protein families database in 2021[J]. Nucleic Acids Res, 2021, 49(D1): D412-D419. |
| [24] | Paysan-Lafosse T, Blum M, Chuguransky S, et al. InterPro in 2022[J]. Nucleic Acids Res, 2023, 51(D1): D418-D427. |
| [25] | Horton P, Park KJ, Obayashi T, et al. WoLF PSORT: protein localization predictor[J]. Nucleic Acids Res, 2007, 35(Web Server issue): W585-W587. |
| [26] |
Arora R, Agarwal P, Ray S, et al. MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress[J]. BMC Genomics, 2007, 8: 242.
pmid: 17640358 |
| [27] | Nguyen LT, Schmidt HA, von Haeseler A, et al. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies[J]. Mol Biol Evol, 2015, 32(1): 268-274. |
| [28] |
Rozas J, Sánchez-DelBarrio JC, Messeguer X, et al. DnaSP, DNA polymorphism analyses by the coalescent and other methods[J]. Bioinformatics, 2003, 19(18): 2496-2497.
doi: 10.1093/bioinformatics/btg359 pmid: 14668244 |
| [29] | Xie JB, Chen SS, Xu WJ, et al. Origination and function of plant Pseudogenes[J]. Plant Signal Behav, 2019, 14(8): 1625698. |
| [30] |
Zou CS, Chen AJ, Xiao LH, et al. A high-quality genome assembly of quinoa provides insights into the molecular basis of salt bladder-based salinity tolerance and the exceptional nutritional value[J]. Cell Res, 2017, 27(11): 1327-1340.
doi: 10.1038/cr.2017.124 pmid: 28994416 |
| [31] | Wu Q, Bai X, Nie MP, et al. Genome-wide identification and expression analysis disclose the pivotal PHOSPHATIDYLETHAN-OLAMINE BINDING PROTEIN members that may be utilized for yield improvement of Chenopodium quinoa[J]. Front Plant Sci, 2022, 13: 1119049. |
| [32] | Mirzaghaderi G. Genome-wide analysis of MADS-box transcription factor gene family in wild emmer wheat(Triticum turgidum subsp. dicoccoides)[J]. PLoS One, 2024, 19(3): e0300159. |
| [33] | Yang XY, Zhang MJ, Xi DX, et al. Genome-wide identification and expression analysis of the MADS gene family in sweet orange(Citrus sinensis)infested with pathogenic bacteria[J]. PeerJ, 2024, 12: e17001. |
| [34] |
Abdullah-Zawawi MR, Ahmad-Nizammuddin NF, Govender N, et al. Comparative genome-wide analysis of WRKY, MADS-box and MYB transcription factor families in Arabidopsis and rice[J]. Sci Rep, 2021, 11(1): 19678.
doi: 10.1038/s41598-021-99206-y pmid: 34608238 |
| [35] | Qiu YC, Li Z, Walther D, et al. Updated phylogeny and protein structure predictions revise the hypothesis on the origin of MADS-box transcription factors in land plants[J]. Mol Biol Evol, 2023, 40(9): msad194. |
| [36] | Jarvis DE, Sproul JS, Navarro-Domínguez B, et al. Chromosome-scale genome assembly of the hexaploid taiwanese goosefoot “djulis”(Chenopodium formosanum)[J]. Genome Biol Evol, 2022, 14(8): evac120. |
| [37] | Li L, Nie MP, Lu J, et al. Genome-wide identification, expression analysis and gene duplication analysis of Dof transcription factors between quinoa and its ancestral diploid sub-genome species reveal key CDF homologs involved in photoperiodic flowering regulation[J]. S Afr N J Bot, 2023, 162(6): 545-558. |
| [38] | Chai SY, Li KX, Deng XX, et al. Genome-wide analysis of the MADS-box gene family and expression analysis during anther development in Salvia miltiorrhiza[J]. Int J Mol Sci, 2023, 24(13): 10937. |
| [39] | Bai G, Yang DH, Cao PJ, et al. Genome-wide identification, gene structure and expression analysis of the MADS-box gene family indicate their function in the development of tobacco(Nicotiana tabacum L.)[J]. Int J Mol Sci, 2019, 20(20): 5043. |
| [40] | Zeng F, Zheng CM, Ge WX, et al. Regulatory function of the endogenous hormone in the germination process of quinoa seeds[J]. Front Plant Sci, 2023, 14: 1322986. |
| [1] | CUI Yuan-yuan, WANG Zhao-yi, BAI Shuang-yu, REN Yu-zhao, DOU Fei-fei, LIU Cai-xia, LIU Feng-lou, WANG Zhang-jun, LI Qing-feng. Genome-wide Identification of Non-specific Phospholipase C Gene Family in Hordeum vulgare L. and Stress Expression Analysis at Seedling Stage [J]. Biotechnology Bulletin, 2024, 40(8): 74-82. |
| [2] | LIN Tong, YUAN Cheng, DONG Chen-wen-hua, ZENG Meng-qiong, YANG Yan, MAO Zi-chao, LIN Chun. Screening and Functional Analysis of Gene CqSTK Associated with Gametophyte Development of Quinoa [J]. Biotechnology Bulletin, 2024, 40(8): 83-94. |
| [3] | ZANG Wen-rui, MA Ming, CHE Gen, HASI Agula. Genome-wide Identification and Expression Pattern Analysis of BZR Transcription Factor Gene Family of Melon [J]. Biotechnology Bulletin, 2024, 40(7): 163-171. |
| [4] | SUN Hui-qiong, ZHANG Chun-lai, WANG Xi-liang, XU Hong-shen, DOU Miao-miao, YANG Bo-hui, CHAI Wen-ting, ZHAO Shan-shan, JIANG Xiao-dong. Identification, Expression and DNA Variation Analysis of FLS Gene Family in Chenopodium quinoa [J]. Biotechnology Bulletin, 2024, 40(7): 172-182. |
| [5] | ALIYA Waili, CHEN Yong-kun, KELAREMU Kelimujiang, WANG Bao-qing, CHEN Ling-na. Phylogenetic Evolution and Expression Analysis of SPL Gene Family in Juglans regia [J]. Biotechnology Bulletin, 2024, 40(6): 180-189. |
| [6] | GONG Li-li, YU Hua, YANG Jie, CHEN Tian-chi, ZHAO Shuang-ying, WU Yue-yan. Identification and Analysis of Grape(Vitis vinifera L.)CYP707A Gene Family and Functional Verification to Fruit Ripening [J]. Biotechnology Bulletin, 2024, 40(2): 160-171. |
| [7] | JIANG Yu-shan, LAN Qian, WANG Fang, JIANG Liang, PEI Cheng-cheng. Characterization of a Quinoa Mutant Affecting Tyrosine Metabolism [J]. Biotechnology Bulletin, 2024, 40(10): 253-261. |
| [8] | NIU De, HE Yue-hui. Molecular Epigenetic Understanding of Seasonal Regulation of Flowering Time in Wheat [J]. Biotechnology Bulletin, 2024, 40(10): 30-40. |
| [9] | ZHANG Lu-yang, HAN Wen-long, XU Xiao-wen, YAO Jian, LI Fang-fang, TIAN Xiao-yuan, ZHANG Zhi-qiang. Identification and Expression Analysis of the Tobacco TCP Gene Family [J]. Biotechnology Bulletin, 2023, 39(6): 248-258. |
| [10] | LAI Rui-lian, FENG Xin, GAO Min-xia, LU Yu-dan, LIU Xiao-chi, WU Ru-jian, CHEN Yi-ting. Genome-wide Identification of Catalase Family Genes and Expression Analysis in Kiwifruit [J]. Biotechnology Bulletin, 2023, 39(4): 136-147. |
| [11] | DUAN Min-jie, LI Yi-fei, YANG Xiao-miao, WANG Chun-ping, HUANG Qi-zhong, HUANG Ren-zhong, ZHANG Shi-cai. Identification of Zinc Finger Protein DnaJ-Like Gene Family in Capsicum annuum and Its Expression Analysis Responses to High Temperature Stress [J]. Biotechnology Bulletin, 2023, 39(1): 187-198. |
| [12] | YUAN Xing, GUO Cai-hua, LIU Jin-ming, KANG Chao, QUAN Shao-wen, NIU Jian-xin. Genome-wide Identification of CONSTANS-Like Family Genes and Expression Analysis in Wanlut [J]. Biotechnology Bulletin, 2022, 38(9): 167-179. |
| [13] | ZHANG Ye-meng, ZHU Li-li, CHEN Zhi-guo. Identification and Expression Analysis of NHX Gene Family in Quinoa Under Salt Stress [J]. Biotechnology Bulletin, 2022, 38(12): 184-193. |
| [14] | CAO Ying-hui, HU Mei-juan, TONG Yan, ZHANG Yan-ping, ZHAO Kai, PENG Dong-hui, ZHOU Yu-zhen. Identification of the ABC Gene Family and Expression Pattern Analysis During Flower Development in Cymbidium ensifolium [J]. Biotechnology Bulletin, 2022, 38(11): 162-174. |
| [15] | GAO Ling, WANG Fei, XIE Shuang-quan, CHEN Xi-feng, SHEN Hai-tao, LI Hong-bin. Genome-wide Identification and Expression Analysis of CBL Gene Family in Glycyrrhiza uralensis [J]. Biotechnology Bulletin, 2021, 37(4): 18-27. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||