








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (10): 121-128.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0383
Previous Articles Next Articles
DUAN Xi-yuan1,2( ), LUO Zhen1, TANG Wei1, LU He-quan1, KONG Xiang-qiang1,2(
), LUO Zhen1, TANG Wei1, LU He-quan1, KONG Xiang-qiang1,2( )
)
Received:2025-04-12
Online:2025-10-26
Published:2025-10-28
Contact:
KONG Xiang-qiang
E-mail:2022020904@stu.sdnu.edu.cn;kongqiang_1995@163.com
DUAN Xi-yuan, LUO Zhen, TANG Wei, LU He-quan, KONG Xiang-qiang. Research Progress in the Mechanism of CEP Regulating Plant Nutrient Uptake[J]. Biotechnology Bulletin, 2025, 41(10): 121-128.
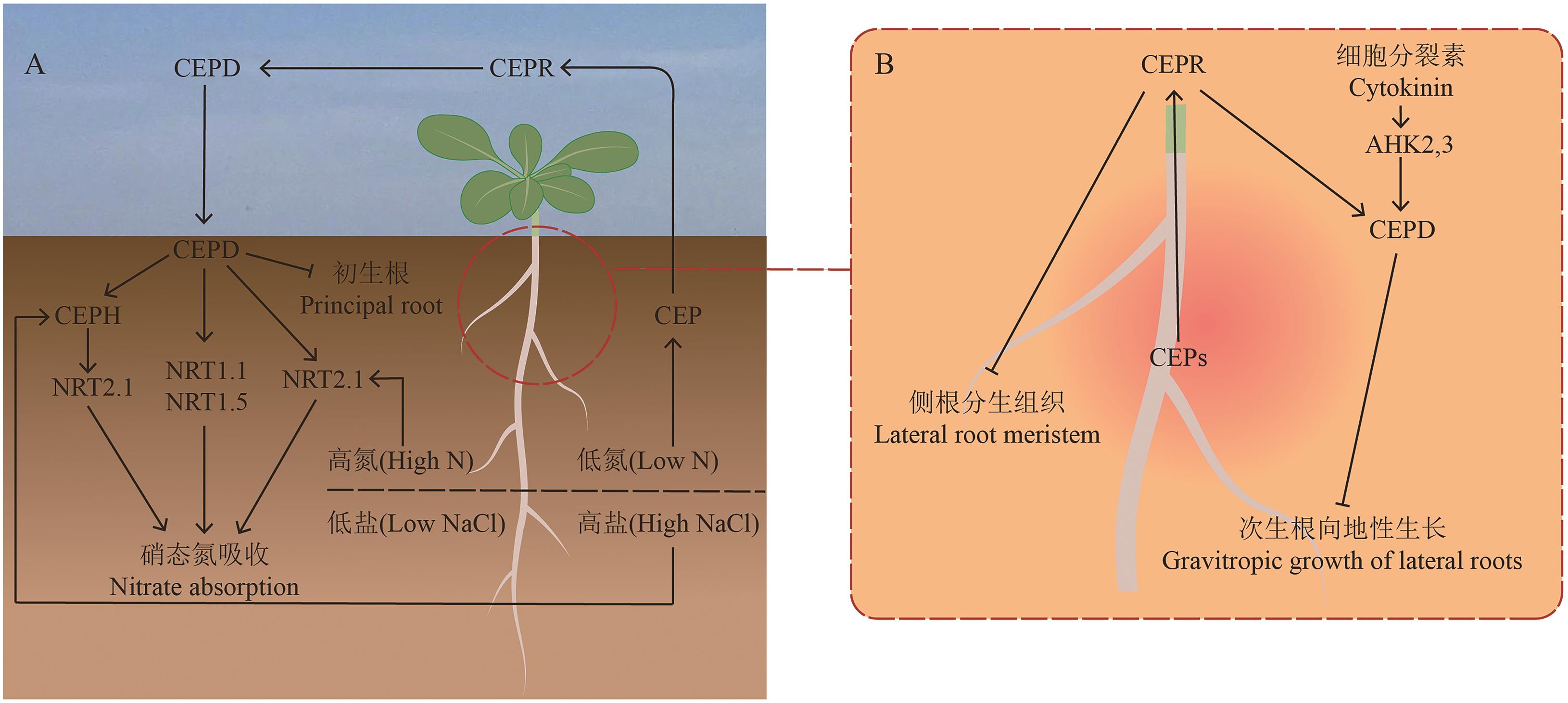
Fig. 1 Mechanism of CEP signaling in regulating nitrate uptake and root growthA: Under heterogeneous nutrient or salt distribution in the root system, low nitrogen (N) or salt stress on one side induces the upregulation of CEP gene expression in the stressed roots. The synthesized CEP peptides are transported to the shoot via the xylem, where they interact with the CEP receptor (CEPR). This interaction triggers the upregulation of CEPD gene expression, leading to the production of CEPD peptides, which are then transported back to the roots via the phloem to exert their effects. First, under high-N/low-salt conditions, CEPD peptides specifically upregulate the transcript encoding NRT2.1, promoting NRT2.1 protein synthesis. Second, CEPD peptides enhance the expressions of NRT1.1 and NRT1.5, thereby improving nitrate uptake and transport. Additionally, CEPH is induced under low-N conditions and further activates NRT2.1 transport activity in response to CEPD. Moreover, CEPD suppresses primary root growth, thus modulating root system architecture. B: The synthesized CEP peptides in the roots, are transported to the shoot. There, they interact with the CEP receptor (CEPR), generating a signal that suppresses lateral root growth. Furthermore, CEPR activates CEPD expression, which inhibits the gravitropic growth of secondary roots. Concurrently, cytokinins promote CEPD expression in the roots via AHK2 and AHK3, increasing CEPD levels in the roots and further suppressing secondary root gravitropism
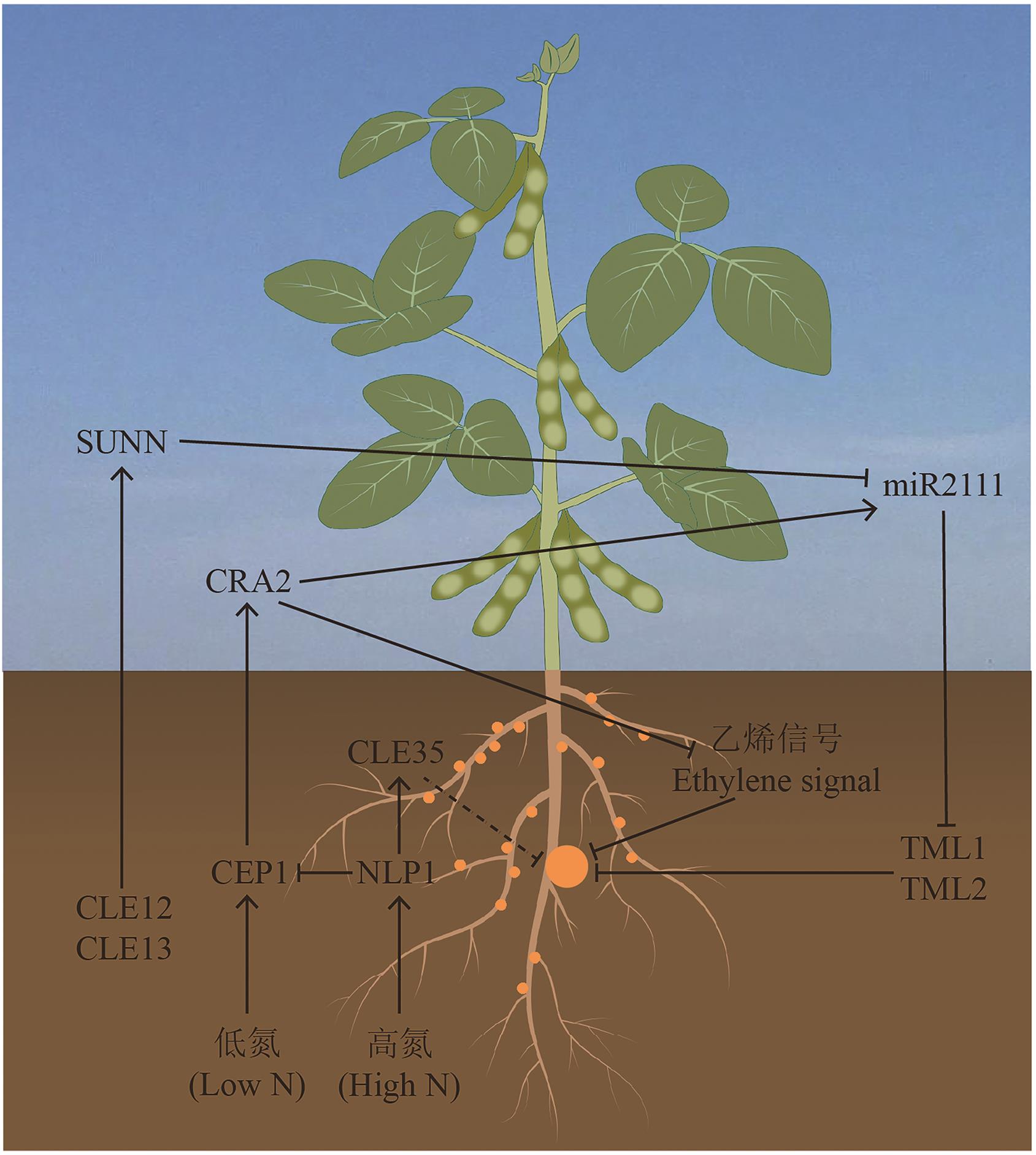
Fig. 2 CEP regulates symbiotic nodulation between plants and rhizobiaIn the regulation of root nodulation, CLEs and CEP family peptides demonstrate antagonistic effects. Under low nitrogen conditions, CEP1 expression is induced in plant roots, and its product is transported to the shoots via the xylem, where it binds to CAR2 to promote miR2111 synthesis and subsequent transport back to the roots, thereby reducing TML1 and TML2 protein accumulation to facilitate nodulation. Meanwhile, CRA2 suppresses ethylene-dependent signaling genes to attenuate the inhibitory effect of ethylene on nodulation. In the autoregulation pathway of nodulation, root-derived CLE12 and CLE13 activate SUNN in the shoots to inhibit miR2111 expression, thereby counteracting the CEP-mediated upregulation of miR2111. When external nitrogen levels are high, NLP1 activates the expression of the negative regulator CLE35 while simultaneously suppress the positive regulator CEP1 to modulate root nodulation
| [1] | Gautrat P, Laffont C, Frugier F, et al. Nitrogen systemic signaling: from symbiotic nodulation to root acquisition [J]. Trends Plant Sci, 2021, 26(4): 392-406. |
| [2] | Delay C, Imin N, Djordjevic MA. CEP genes regulate root and shoot development in response to environmental cues and are specific to seed plants [J]. J Exp Bot, 2013, 64(17): 5383-5394. |
| [3] | Imin N, Mohd-Radzman NA, Ogilvie HA, et al. The peptide-encoding CEP1 gene modulates lateral root and nodule numbers in Medicago truncatula [J]. J Exp Bot, 2013, 64(17): 5395-5409. |
| [4] | Tabata R, Sumida K, Yoshii T, et al. Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling [J]. Science, 2014, 346(6207): 343-346. |
| [5] | Chapman K, Taleski M, Ogilvie HA, et al. CEP-CEPR1 signalling inhibits the sucrose-dependent enhancement of lateral root growth [J]. J Exp Bot, 2019, 70(15): 3955-3967. |
| [6] | Pan X, Deng Z, Wu R, et al. Identification of CEP peptides encoded by the tobacco (Nicotiana tabacum) genome and characterization of their roles in osmotic and salt stress responses [J]. Plant Physiol Biochem, 2024, 209: 108525. |
| [7] | Patel N, Mohd-Radzman NA, Corcilius L, et al. Diverse peptide hormones affecting root growth identified in the Medicago truncatula secreted peptidome [J]. Mol Cell Proteomics, 2018, 17(1): 160-174. |
| [8] | Huang Y, Ji Z, Zhang S, et al. Function of hormone signaling in regulating nitrogen-use efficiency in plants [J]. J Plant Physiol, 2024, 294: 154191. |
| [9] | Jia Z, Giehl RFH, Von Wirén N. The root foraging response under low nitrogen depends on DWARF1-mediated brassinosteroid biosynthesis [J]. Plant Physiol, 2020, 183(3): 998-1010. |
| [10] | Ohkubo Y, Tanaka M, Tabata R, et al. Shoot-to-root mobile polypeptides involved in systemic regulation of nitrogen acquisition [J]. Nat Plants, 2017, 3: 17029. |
| [11] | Li C, Hu Q, Luo Z, et al. C-terminally encoded peptides act as signals to increase cotton root nitrate uptake under nonuniform salinity [J]. Plant Physiol, 2023, 194(1): 530-545. |
| [12] | Ohkubo Y, Kuwata K, Matsubayashi Y. A type 2C protein phosphatase activates high-affinity nitrate uptake by dephosphorylating NRT2.1 [J]. Nat Plants, 2021, 7(3): 310-316. |
| [13] | Furumizu C, Aalen RB. Peptide signaling through leucine-rich repeat receptor kinases: insight into land plant evolution [J]. New Phytol, 2023, 238(3): 977-982. |
| [14] | Luo Z, Moreau C, Wang J, et al. NLP1 binds the CEP1 signalling peptide promoter to repress its expression in response to nitrate [J]. New Phytol, 2022, 234(5): 1547-1552. |
| [15] | Luo Z, Wang J, Li F, et al. The small peptide CEP1 and the NIN-like protein NLP1 regulate NRT2.1 to mediate root nodule formation across nitrate concentrations [J]. Plant Cell, 2023, 35(2): 776-794. |
| [16] | Taleski M, Jin M, Chapman K, et al. CEP hormones at the nexus of nutrient acquisition and allocation, root development, and plant-microbe interactions [J]. J Exp Bot, 2024, 75(2): 538-552. |
| [17] | Sui Z, Wang T, Li H, et al. Overexpression of peptide-encoding OsCEP6.1 results in pleiotropic effects on growth in rice (O. sativa) [J]. Front Plant Sci, 2016, 7: 228. |
| [18] | Xu R, Li Y, Sui Z, et al. A C-terminal encoded peptide, ZmCEP1, is essential for kernel development in maize [J]. J Exp Bot, 2021, 72(15): 5390-5406. |
| [19] | Ogilvie H A, Imin N, Djordjevic MA. Diversification of the C-TERMINALLY ENCODED PEPTIDE (CEP) gene family in angiosperms, and evolution of plant-family specific CEP genes [J]. BMC Genomics, 2014, 15(1): 870. |
| [20] | Lu F, Wang X, Liu B, et al. A C-Terminally Encoded Peptide, MeCEP6, promotes nitrate uptake in cassava roots [J]. Plants (Basel), 2025, 14(8). |
| [21] | Chu X, Li M, Zhang S, et al. HBI1-TCP20 interaction positively regulates the CEPs-mediated systemic nitrate acquisition [J]. J Integr Plant Biol, 2021, 63(5): 902-912. |
| [22] | Jacquot A, Chaput V, Mauries A, et al. NRT2.1 C-terminus phosphorylation prevents root high affinity nitrate uptake activity in Arabidopsis thaliana [J]. New Phytol, 2020, 228(3): 1038-1054. |
| [23] | Zhang L, Yu Z, Xu Y, et al. Regulation of the stability and ABA import activity of NRT1.2/NPF4.6 by CEPR2-mediated phosphorylation in Arabidopsis [J]. Mol Plant, 2022, 15(10): 1635. |
| [24] | Roy S, Griffiths M, Torres-Jerez I, et al. Application of Synthetic Peptide CEP1 increases nutrient uptake rates along plant roots [J]. Front Plant Sci, 2021, 12: 793145. |
| [25] | Kawai M, Tabata R, Ohashi M, et al. Regulation of ammonium acquisition and use in Oryza longistaminata ramets under nitrogen source heterogeneity [J]. Plant Physiol, 2022, 188(4): 2364-2376. |
| [26] | Delay C, Chapman K, Taleski M, et al. CEP3 levels affect starvation-related growth responses of the primary root [J]. J Exp Bot, 2019, 70(18): 4763-4774. |
| [27] | Yu Z, Qu X, Lv B, et al. MAC3A and MAC3B mediate degradation of the transcription factor ERF13 and thus promote lateral root emergence [J]. Plant Cell, 2024, 36(9): 3162-3176. |
| [28] | Kiba T, Krapp A. Plant nitrogen acquisition under low availability: Regulation of uptake and root architecture [J]. Plant Cell Physiol, 2016, 57(4): 707-714. |
| [29] | Wang N Q, Kong C H, Wang P, et al. Root exudate signals in plant-plant interactions [J]. Plant Cell Environ, 2021, 44(4): 1044-1058. |
| [30] | Oh E, Seo P J, Kim J. Signaling peptides and receptors coordinating plant root development [J]. Trends Plant Sci, 2018, 23(4): 337-351. |
| [31] | Lu S, Xiao F. Small peptides: orchestrators of plant growth and developmental processes [J]. Int J Mol Sci, 2024, 25(14). |
| [32] | Mei Z, Li B, Zhu S, et al. A genome-wide analysis of the CEP gene family in cotton and a functional study of GhCEP46-D05 in plant development [J]. Int J Mol Sci, 2024, 25(8). |
| [33] | Li X, Cai W, Liu Y, et al. Differential TOR activation and cell proliferation in Arabidopsis root and shoot apexes [J]. Proc Natl Acad Sci U S A, 2017, 114(10): 2765-2770. |
| [34] | Mohd-Radzman N A, Laffont C, Ivanovici A, et al. Different pathways act downstream of the CEP Peptide Receptor CRA2 to regulate lateral root and nodule development [J]. Plant Physiol, 2016, 171(4): 2536-2548. |
| [35] | Baena-González E, Rolland F, Thevelein JM, et al. A central integrator of transcription networks in plant stress and energy signalling [J]. Nature, 2007, 448(7156): 938-942. |
| [36] | Huang A, Cui T, Zhang Y, et al. CRISPR/Cas9-engineered large fragment deletion mutations in Arabidopsis CEP peptide-encoding genes reveal their role in primary and lateral root formation [J]. Plant Cell Physiol, 2023, 64(1): 19-26. |
| [37] | Zhu F, Ye Q, Chen H, et al. Multigene editing reveals that MtCEP1/2/12 redundantly control lateral root and nodule number in Medicago truncatula [J]. J Exp Bot, 2021, 72(10): 3661-3676. |
| [38] | Roychoudhry S, Kieffer M, Del Bianco M, et al. The developmental and environmental regulation of gravitropic setpoint angle in Arabidopsis and bean [J]. Sci Rep, 2017, 7: 42664. |
| [39] | Chapman K, Ivanovici A, Taleski M, et al. CEP receptor signalling controls root system architecture in Arabidopsis and Medicago [J]. New Phytol, 2020, 226(6): 1809-1821. |
| [40] | Chapman K, Taleski M, Frank M, et al. C-TERMINALLY ENCODED PEPTIDE (CEP) and cytokinin hormone signaling intersect to promote shallow lateral root angles [J]. J Exp Bot, 2024, 75(2): 631-641. |
| [41] | Chang L, Ramireddy E, Schmülling T. Lateral root formation and growth of Arabidopsis is redundantly regulated by cytokinin metabolism and signalling genes [J]. J Exp Bot, 2013, 64(16): 5021-5032. |
| [42] | Lin J, Frank M, Reid D. No home without hormones: How plant hormones control legume nodule organogenesis [J]. Plant Commun, 2020, 1(5): 100104. |
| [43] | Taleski M, Chapman K, Novák O, et al. CEP peptide and cytokinin pathways converge on CEPD glutaredoxins to inhibit root growth [J]. Nat Commun, 2023, 14(1): 1683. |
| [44] | Ota R, Ohkubo Y, Yamashita Y, et al. Shoot-to-root mobile CEPD-like 2 integrates shoot nitrogen status to systemically regulate nitrate uptake in Arabidopsis [J]. Nat Commun, 2020, 11(1): 641. |
| [45] | Laffont C, Frugier F. Rhizobium symbiotic efficiency meets CEP signaling peptides [J]. New Phytol, 2024, 241(1): 24-27. |
| [46] | Roy S, Liu W, Nandety RS, et al. Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation [J]. Plant Cell, 2020, 32(1): 15-41. |
| [47] | Cervantes-Pérez SA, Zogli P, Amini S, et al. Single-cell transcriptome atlases of soybean root and mature nodule reveal new regulatory programs that control the nodulation process [J]. Plant Commun, 2024, 5(8): 100984. |
| [48] | Soyano T, Akamatsu A, Takeda N, et al. Periodic cytokinin responses in Lotus japonicus rhizobium infection and nodule development [J]. Science, 2024, 385(6706): 288-294. |
| [49] | Lin J, Bjørk P K, Kolte MV, et al. Zinc mediates control of nitrogen fixation via transcription factor filamentation [J]. Nature, 2024, 631(8019): 164-169. |
| [50] | Ivanovici A, Laffont C, Larrainzar E, et al. The medicago SymCEP7 hormone increases nodule number via shoots without compromising lateral root number [J]. Plant Physiol, 2023, 191(3): 2012-2026. |
| [51] | Tsikou D, Yan Z, Holt DB, et al. Systemic control of legume susceptibility to rhizobial infection by a mobile microRNA [J]. Science, 2018, 362(6411): 233-236. |
| [52] | Penmetsa RV, Uribe P, Anderson J, et al. The Medicago truncatula ortholog of Arabidopsis EIN2, sickle, is a negative regulator of symbiotic and pathogenic microbial associations [J]. Plant J, 2008, 55(4): 580-595. |
| [53] | Cui S, Inaba S, Suzaki T, et al. Developing for nutrient uptake: Induced organogenesis in parasitic plants and root nodule symbiosis [J]. Curr Opin Plant Biol, 2023, 76: 102473. |
| [54] | Zhu F, Deng J, Chen H, et al. A CEP peptide receptor-like kinase regulates auxin biosynthesis and ethylene signaling to coordinate root growth and symbiotic nodulation in Medicago truncatula [J]. Plant Cell, 2020, 32(9): 2855-2877. |
| [55] | Laffont C, Huault E, Gautrat P, et al. Independent regulation of symbiotic nodulation by the SUNN negative and CRA2 positive systemic pathways [J]. Plant Physiol, 2019, 180(1): 559-570. |
| [56] | Hazak O, Hardtke CS. CLAVATA 1-type receptors in plant development [J]. J Exp Bot, 2016, 67(16): 4827-4833. |
| [57] | Hussain S, Wang W, Ahmed S, et al. PIP2, an auxin induced plant peptide hormone regulates root and hypocotyl elongation in Arabidopsis [J]. Front Plant Sci, 2021, 12: 646736. |
| [58] | Schnabel E, Journet EP, De Carvalho-Niebel F, et al. The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length [J]. Plant Mol Biol, 2005, 58(6): 809-822. |
| [59] | Laffont C, Ivanovici A, Gautrat P, et al. The NIN transcription factor coordinates CEP and CLE signaling peptides that regulate nodulation antagonistically [J]. Nat Commun, 2020, 11(1): 3167. |
| [60] | Gautrat P, Laffont C, Frugier F. Compact root architecture 2 promotes root competence for nodulation through the miR2111 systemic effector [J]. Curr Biol, 2020, 30(7): 1339-1345.e1333. |
| [61] | Imin N, Patel N, Corcilius L, et al. CLE peptide tri-arabinosylation and peptide domain sequence composition are essential for SUNN-dependent autoregulation of nodulation in Medicago truncatula [J]. New Phytol, 2018, 218(1): 73-80. |
| [62] | Luo Z, Tang W, Wang XW, et al. Effects of N application methods on cotton yield and fertilizer N recovery efficiency in salinity fields with drip irrigation under mulch film using 15N [J]. Front Plant Sci, 2024, 15:1394285. |
| [1] | KANG Qin, WANG Xia, SHEN Ming-yang, XU Jing-tian, CHEN Shi-lan, LIAO Ping-yang, XU Wen-zhi, WU Wei, XU Dong-bei. Cloning and Expression Analysis of the UV-B Receptor Gene McUVR8 in Mentha canadensis L. [J]. Biotechnology Bulletin, 2025, 41(8): 255-266. |
| [2] | LI Si-bo, QIAN Hong-ping, XU Chang-wen, WANG Xiao, LIN Jin-xing, CUI Ya-ning. Research Progress in the Involvement of Intracellular Transport Regulated by Endogenous Elicitors in Plant Growth and Development and Response to Adverse Stress [J]. Biotechnology Bulletin, 2025, 41(7): 17-27. |
| [3] | LIU Li, WANG Hui, GUAN Tian-shu, LI Bai-hong, YU Shu-yi. Screening the Interacting Protein of Abscisic Acid Receptor VvPYL4 and the Gene Expression of the Interacting Protein in Grape [J]. Biotechnology Bulletin, 2025, 41(4): 188-197. |
| [4] | LI Wen-cui, PENG Yu-jia, LIU Yong-bo. Research Progress in Light Signal and Circadian Rhythm Regulating the Perception of Plants to Cold Stress [J]. Biotechnology Bulletin, 2024, 40(8): 1-12. |
| [5] | YAN Huan-huan, SHANG Yi-tong, WANG Li-hong, TIAN Xue-qin, LIAO Hai-yan, ZENG Bin, HU Zhi-hong. Heterologous Biosynthesis of Cordycepin in Aspergillus oryzae [J]. Biotechnology Bulletin, 2024, 40(6): 290-298. |
| [6] | XIE Yan-jie. Harmony in Diversity: Auxin and Redox Signaling During Root Hair Development [J]. Biotechnology Bulletin, 2024, 40(6): 1-4. |
| [7] | CHEN Yan-mei. Crosstalk Between Different Post-translational Modifications and Its Regulatory Mechanisms in Plant Growth and Development [J]. Biotechnology Bulletin, 2024, 40(2): 1-8. |
| [8] | JI Zhong-xiang, LUORENG Zhuo-ma, LI Yu-hang, WANG Yu-mei, HU Xi-min, LI Yan-qing, WANG Xing-ping. Roles of miR-3604 on the Receptivity, Proliferation, and Apoptosis of Bovine Endometrial Epithelial Cells [J]. Biotechnology Bulletin, 2024, 40(12): 291-298. |
| [9] | LI Qing, SHI Yu-he, ZHU Jue, LI Xiao-ling, HOU Chao-wen, TONG Qiao-zhen. Genetic Diversity Analysis and DNA Fingerprint Construction of Atractylodes macrocephala Germplasm Resources Based on SCoT Molecular Markers [J]. Biotechnology Bulletin, 2024, 40(11): 142-151. |
| [10] | XIONG Xin-yi, LIU Li-ping, FENG Jie, ZHANG Jin-song, LI De-shun, LIU Peng, LIU Yan-fang. Effect and Mechanism of Cordyceps militaris Extract on Lowering Uric Acid in Hyperuricemia Rats [J]. Biotechnology Bulletin, 2024, 40(11): 34-46. |
| [11] | CHEN Meng-jiao, LI Yang-yang, WU Qian. Research Advances in Plant Growth and Stress Response Regulation Mediated by Glutamate Receptor-like Proteins [J]. Biotechnology Bulletin, 2024, 40(10): 62-75. |
| [12] | HU Hai-lin, XU Li, LI Xiao-xu, WANG Chen-can, MEI Man, DING Wen-jing, ZHAO Yuan-yuan. Advances in the Regulation of Plant Growth, Development and Stress Physiology by Small Peptide Hormones [J]. Biotechnology Bulletin, 2023, 39(7): 13-25. |
| [13] | LI Tuo, LI Long-ping, QU Lei. Research Progress in the Structure of Tailed Bacteriophage and Its Receptors [J]. Biotechnology Bulletin, 2023, 39(6): 88-101. |
| [14] | SUN Ya-ling, LI Rui-ping, WANG Zhen-bao, ZHANG Shu, LIU Bing-jiang, HUO Yu-meng. A New Method for Onion Seed Disinfection and Aseptic Seedling Culture [J]. Biotechnology Bulletin, 2023, 39(4): 212-220. |
| [15] | YAO Xiao-wen, LIANG Xiao, CHEN Qing, WU Chun-ling, LIU Ying, LIU Xiao-qiang, SHUI Jun, QIAO Yang, MAO Yi-ming, CHEN Yin-hua, ZHANG Yin-dong. Study on the Expression Pattern of Genes in Lignin Biosynthesis Pathway of Cassava Resisting to Tetranychus urticae [J]. Biotechnology Bulletin, 2023, 39(2): 161-171. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||