








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (10): 156-163.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0662
Previous Articles Next Articles
YANG Yang( ), LIU Hui-min, LIN Li, WANG You-ping, WU Jian(
), LIU Hui-min, LIN Li, WANG You-ping, WU Jian( )
)
Received:2025-06-23
Online:2025-10-26
Published:2025-10-28
Contact:
WU Jian
E-mail:2923977275@qq.com;wu_jian@yzu.edu.cn
YANG Yang, LIU Hui-min, LIN Li, WANG You-ping, WU Jian. A High-throughput and Rapid Method for Plant Genomic DNA Extraction[J]. Biotechnology Bulletin, 2025, 41(10): 156-163.
引物名称 Primer name | 正向引物序列 Forward primer sequence (5′‒3′) | 反向引物序列 Reverse primer sequence (5′‒3′) | 产物大小 Amplicon size (bp) |
|---|---|---|---|
| BnaC06.IDA | CAAACCAGATTCCCATTTCC | GGAATGGGAACACCTTTGGG | 514 |
| BnaA07.IDA | AAGCGAAAAGATAAGCTTGC | CGGATGATGTGTGATGCTGGA | 974 |
| ProBnaC06.IDA | GGGTGAACCCAAGAATAAC | TCGCCGCCAGAAACAGAACC | 1 215 |
| ProBnaA07.IDA | CTTGGGGCAACCCTAATCTG | CACGGAGCCATTTGGTAAT | 1 634 |
| ProBnaC04.IDA | CCACCACAAACGAGAAGACG | CACGGAGCCATTTGGTAATG | 1 803 |
| OsActin | GGTGTCATGGTCGGAATGGG | GTCCAGGGCGATGTAGGAAA | 792 |
| InDel205 | TTAAGCCCAAAACAAATTCAAATGGACTAAAA | CAAAATTTGGTTGCATAACCCTATTTCTCTTT | 127 |
| InDel348 | GAAAATAAAATTGGGAGTGGGAGGGAAA | TTTTCTCTAACGTCTCCACACGAAGATTAT | 130 |
| InDel299 | GAAACTAACTAATTTCACTTTCTCGCTCATCT | CTTTGATACTGTTGAACTAAACCGACTTTTCT | 101 |
| InDel322 | CACCCACAAACCAAAAACTAAAATTATACGAA | GCTGTAGTCTCGGTATCTCAGATTGTG | 116 |
| InDel204 | GAAGAAGCTTCGAATTTGACGGTGAC | GTCATTATTTTATGTATTTCATCCCGGGCG | 108 |
| InDel295 | AAAGCTGGCACGTCACAATATCAAT | CGAGCTATCTTTTGTTTTGGTGATATATCCTT | 130 |
| InDel319 | AGTTTTGAGTCGTATCTGAAATAAACAAAAGA | AGCACTACTTGTCGGAAAATAAAATAACATGA | 130 |
| InDel285 | TAAAAATCATTATGCAGACCGTAACTGGTAAT | ATGATGAAAATCTAAAGTGTTTTCCGTCAAAG | 106 |
| InDel620 | AGGAAAACCCCCAAAAGTAAATTTAAAAACAT | TATTGATTTTCCTCCACTGATTTGGTTTTCTA | 104 |
| InDel599 | CCATTGTAAACATCCTACGTATTGATTACTGA | CAAGCTTGTGAAAAGAAAGAAGAAGAAAATGT | 116 |
| InDel605 | GACTGATACCCACATGTATTAGAAGCAATGAC | GACTTTTACCCGGAACTGTTTACTTGAA | 119 |
Table 1 Primer sequences used in this study
引物名称 Primer name | 正向引物序列 Forward primer sequence (5′‒3′) | 反向引物序列 Reverse primer sequence (5′‒3′) | 产物大小 Amplicon size (bp) |
|---|---|---|---|
| BnaC06.IDA | CAAACCAGATTCCCATTTCC | GGAATGGGAACACCTTTGGG | 514 |
| BnaA07.IDA | AAGCGAAAAGATAAGCTTGC | CGGATGATGTGTGATGCTGGA | 974 |
| ProBnaC06.IDA | GGGTGAACCCAAGAATAAC | TCGCCGCCAGAAACAGAACC | 1 215 |
| ProBnaA07.IDA | CTTGGGGCAACCCTAATCTG | CACGGAGCCATTTGGTAAT | 1 634 |
| ProBnaC04.IDA | CCACCACAAACGAGAAGACG | CACGGAGCCATTTGGTAATG | 1 803 |
| OsActin | GGTGTCATGGTCGGAATGGG | GTCCAGGGCGATGTAGGAAA | 792 |
| InDel205 | TTAAGCCCAAAACAAATTCAAATGGACTAAAA | CAAAATTTGGTTGCATAACCCTATTTCTCTTT | 127 |
| InDel348 | GAAAATAAAATTGGGAGTGGGAGGGAAA | TTTTCTCTAACGTCTCCACACGAAGATTAT | 130 |
| InDel299 | GAAACTAACTAATTTCACTTTCTCGCTCATCT | CTTTGATACTGTTGAACTAAACCGACTTTTCT | 101 |
| InDel322 | CACCCACAAACCAAAAACTAAAATTATACGAA | GCTGTAGTCTCGGTATCTCAGATTGTG | 116 |
| InDel204 | GAAGAAGCTTCGAATTTGACGGTGAC | GTCATTATTTTATGTATTTCATCCCGGGCG | 108 |
| InDel295 | AAAGCTGGCACGTCACAATATCAAT | CGAGCTATCTTTTGTTTTGGTGATATATCCTT | 130 |
| InDel319 | AGTTTTGAGTCGTATCTGAAATAAACAAAAGA | AGCACTACTTGTCGGAAAATAAAATAACATGA | 130 |
| InDel285 | TAAAAATCATTATGCAGACCGTAACTGGTAAT | ATGATGAAAATCTAAAGTGTTTTCCGTCAAAG | 106 |
| InDel620 | AGGAAAACCCCCAAAAGTAAATTTAAAAACAT | TATTGATTTTCCTCCACTGATTTGGTTTTCTA | 104 |
| InDel599 | CCATTGTAAACATCCTACGTATTGATTACTGA | CAAGCTTGTGAAAAGAAAGAAGAAGAAAATGT | 116 |
| InDel605 | GACTGATACCCACATGTATTAGAAGCAATGAC | GACTTTTACCCGGAACTGTTTACTTGAA | 119 |
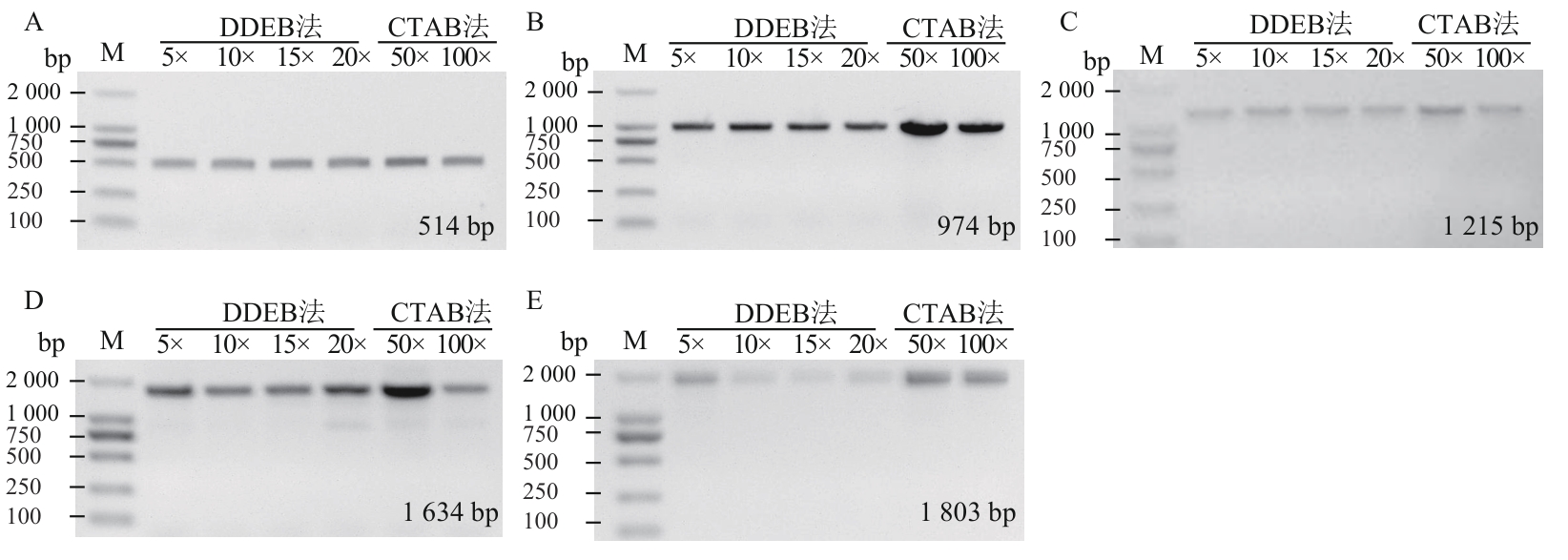
Fig. 2 Comparative PCR amplification efficiency of rapeseed genomic DNA extracted by DDEB and CTAB methods across varying dilution gradientsA‒E the amplified fragment sizes are 514 bp (A), 974 bp (B), 1 215 bp (C), 1 634 bp (D), and 1 803 bp (E), respectively; M: DNA marker
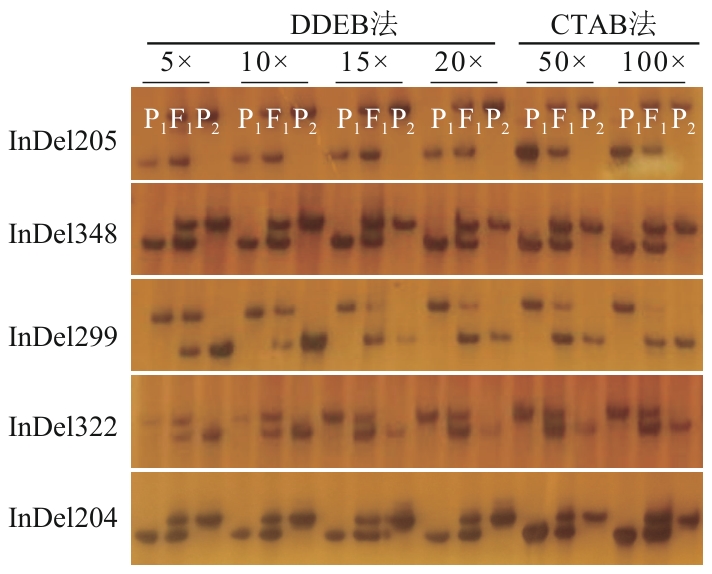
Fig. 4 Comparative evaluation of DDEB and CTAB methods for rapeseed genomic DNA preparation in InDel marker analysisP1: Wanyou 29; P2: SWU66; F1: the hybrid of 'Wanyou 29' (female parent)×'SWU66' (male parent)
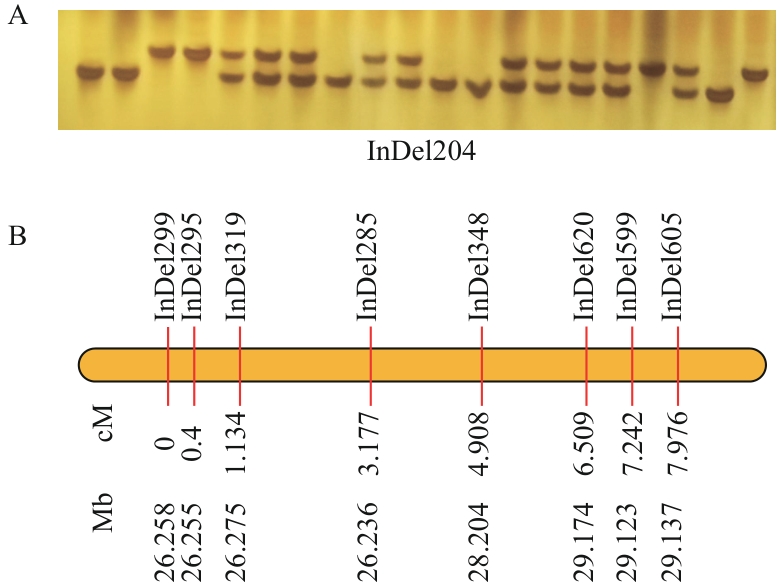
Fig. 5 Analysis of partial genetic linkage map constructed using rapeseed genomic DNA extracted by the DDEB methodA: Genotyping results of InDel204 marker in F₂ population (polyacrylamide gel electrophoresis image). B: Partial genetic linkage map showing (from top to bottom): marker names, genetic distances (cM), and corresponding physical positions (Mb)
| [1] | Amiteye S. Basic concepts and methodologies of DNA marker systems in plant molecular breeding [J]. Heliyon, 2021, 7(10): e08093. |
| [2] | Su XM, Lyu HJ, Li J, et al. Map-based cloning and characterization of yg-2, a gene conferring yellow-green leaf in tomato (Solanum lycopersicum) [J]. Mol Breed, 2024, 44(12): 81. |
| [3] | Wang JC, Chen YJ, Zou Q. Comparative genomics and functional genomics analysis in plants [J]. Int J Mol Sci, 2023, 24(7): 6539. |
| [4] | Ben-Amar A, Mliki A. Timely gene detection assay and reliable screening of genetically engineered plants using an improved direct PCR-based technology [J]. Transgenic Res, 2021, 30(3): 263-274. |
| [5] | Holst-Jensen A, Rønning SB, Løvseth A, et al. PCR technology for screening and quantification of genetically modified organisms (GMOs) [J]. Anal Bioanal Chem, 2003, 375(8): 985-993. |
| [6] | Avenot HF, Jaime-Frias R, Travadon R, et al. Development of PCR-based assays for rapid and reliable detection and identification of canker-causing pathogens from symptomatic almond trees [J]. Phytopathology, 2022, 112(8): 1710-1722. |
| [7] | Komori T, Nitta N. A simple method to control the seed purity of Japonica hybrid rice varieties using PCR-based markers [J]. Plant Breed, 2004, 123(6): 549-553. |
| [8] | Stewart CN Jr, Via LE. A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications [J]. Biotechniques, 1993, 14(5): 748-750. |
| [9] | Porebski S, Bailey LG, Baum BR. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components[J]. Plant Mol Biol Rep, 1997, 15(1): 8-15. |
| [10] | Allen GC, Flores-Vergara MA, Krasynanski S, et al. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide [J]. Nat Protoc, 2006, 1(5): 2320-2325. |
| [11] | Feliciello I, Chinali G. A modified alkaline lysis method for the preparation of highly purified plasmid DNA from Escherichia coli [J]. Anal Biochem, 1993, 212(2): 394-401. |
| [12] | Xia YM, Chen FS, Du Y, et al. A modified SDS-based DNA extraction method from raw soybean [J]. Biosci Rep, 2019, 39(2): BSR20182271. |
| [13] | Werner O, Ros RM, Guerra J. Direct amplification and NaOH extraction: two rapid and simple methods for preparing bryophyte DNA for polymerase chain reaction (PCR) [J]. J Bryol, 2002, 24(2): 127-131. |
| [14] | Putri ND, Prayekti E. Optimalization of NaOH concentration in alkaline lysis method on quality and quantity of Candida albicans DNA [J]. Journal of Indonesian Medical Laboratory and Science, 2024, 5(2): 78-87. |
| [15] | 赵金艳, 刘榜, 王文君, 等. 一种适于PCR扩增的快速提取猪基因组DNA的碱裂解法 [J]. 华中农业大学学报, 2017, 36(4): 90-94. |
| Zhao JY, Liu B, Wang WJ, et al. A rapid alkaline lysis method of extracting pig genomic DNA for PCR amplification [J]. J Huazhong Agric Univ, 2017, 36(4): 90-94. | |
| [16] | 曹文波, 郑璐璐, 谢文海. 一种提取植物基因组DNA的方法——改良尿素法 [J]. 华中师范大学学报: 自然科学版, 2008, 42(3): 448-451. |
| Cao WB, Zheng LL, Xie WH. The modification urea method: an improved method for plant DNA isolation [J]. J Huazhong Norm Univ Nat Sci, 2008, 42(3): 448-451. | |
| [17] | 穆春华, 张发军, 李文才, 等. 玉米叶片基因组快速提取方法研究 [J]. 玉米科学, 2010, 18(3): 170-172. |
| Mu CH, Zhang FJ, Li WC, et al. A method of genomic DNA extraction of maize [J]. J Maize Sci, 2010, 18(3): 170-172. | |
| [18] | 肖帅, 李鳞霞, 李海鸥. 一种高效的植物DNA提取和PCR扩增体系建立 [J]. 亚热带植物科学, 2018, 47(3): 207-210. |
| Xiao S, Li LX, Li HO. An efficient plant DNA extraction and PCR amplification system [J]. Subtrop Plant Sci, 2018, 47(3): 207-210. | |
| [19] | 陈欲, 汪小福, 陈笑芸, 等. 基因组DNA快速提取及在转基因大豆检测中的应用 [J]. 中国油料作物学报, 2021, 43(1): 70-76. |
| Chen Y, Wang XF, Chen XY, et al. Rapid extraction of genomic DNA and its application in detection of genetically modified soybeans [J]. Chin J Oil Crop Sci, 2021, 43(1): 70-76. | |
| [20] | 金素奎, 国倩倩, 刘巧泉, 等. 一种水稻叶片基因组DNA简易提取方法 [J]. 生物技术通报, 2025, 41(1): 74-84. |
| Jin SK, Guo QQ, Liu QQ, et al. A simplified method for extracting genomic DNA from rice leaves [J]. Biotechnol Bull, 2025, 41(1): 74-84. | |
| [21] | 王慧娜, 初志战, 马兴亮, 等. 高通量PCR模板植物基因组DNA制备方法 [J]. 作物学报, 2013, 39(7): 1200-1205. |
| Wang HN, Chu ZZ, Ma XL, et al. A high through-put protocol of plant genomic DNA preparation for PCR [J]. Acta Agron Sin, 2013, 39(7): 1200-1205. | |
| [22] | Ahmed I, Islam M, Arshad W, et al. High-quality plant DNA extraction for PCR: an easy approach [J]. J Appl Genet, 2009, 50(2): 105-107. |
| [23] | Bellstedt DU, Pirie MD, Visser JC, et al. A rapid and inexpensive method for the direct PCR amplification of DNA from plants [J]. Am J Bot, 2010, 97(7): e65-e68. |
| [24] | Hwang H, Bae SC, Lee S, et al. A rapid and simple genotyping method for various plants by direct-PCR [J]. Plant Breed Biotech, 2013, 1(3): 290-297. |
| [25] | Sajib AA, Bhuiya MAI, Huque R. A simple, efficient and rapid method for good quality DNA extraction from rice grains [J]. Rice Sci, 2017, 24(2): 119-122. |
| [26] | Wang TY, Wang L, Zhang JH, et al. A simplified universal genomic DNA extraction protocol suitable for PCR [J]. Genet Mol Res, 2011, 10(1): 519-525. |
| [27] | Li X, Liu C, Wang DB, et al. Persistent pollution of genetic materials in a typical laboratory environment [J]. J Hazard Mater, 2024, 470: 134201. |
| [28] | Wu YH, Bhat PR, Close TJ, et al. Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of a graph [J]. PLoS Genet, 2008, 4(10): e1000212. |
| [29] | Wang BW, Liu XL, Li ZX, et al. A nuclease-dead Cas9-derived tool represses target gene expression [J]. Plant Physiol, 2024, 195(3): 1880-1892. |
| [30] | Abd Razak WNR, Bahrain KBS, Bostanudin MF, et al. Evaluation of the inhibitory effects of pharmaceutical excipients toward the efficiency of polymerase chain reaction (PCR) [J]. Int J Pharm Res, 2020, 12(3): 1968-1976. |
| [1] | LIU Jia, REN Yi-shang, JING Xiao-yan, XU La. Applications and Prospects of Raman Spectroscopy-based “Screen-First, Culture-Second” Strategy in Functional Microbial Resource Exploration [J]. Biotechnology Bulletin, 2026, 42(5): 1-13. |
| [2] | WANG Fang, SHAO Hui-ru, LYU Lin-long, ZHAO Dian, HU Zhen, LYU Jian-zhen, JIANG Liang. Establishment of TurboID Proximity Labeling Technology in Plants and Bacteria [J]. Biotechnology Bulletin, 2025, 41(9): 44-53. |
| [3] | DENG Mei-bi, YAN Lang, ZHAN Zhi-tian, ZHU Min, HE Yu-bing. Efficient CRISPR Gene Editing in Rice Assisted by RUBY [J]. Biotechnology Bulletin, 2025, 41(8): 65-73. |
| [4] | HOU Ya-tao, LI Ying-hui, DENG Lei, LI Chang-bao, LI Chuan-you, SUN Chuan-long. Development of Functional Molecular Markers for Tomato Fruit Weight Gene and Population Genotyping Analysis [J]. Biotechnology Bulletin, 2025, 41(4): 98-105. |
| [5] | ZHOU Di, WANG Dong-xu, GE Sangquzhen, OU Mei-xiang, GUO Xiao-fang, DE Ji. Fungal Diversity, Community Structure and Prediction of Ecological Function in Basomtso Lake, Xizang [J]. Biotechnology Bulletin, 2025, 41(1): 298-311. |
| [6] | JIN Su-kui, GUO Qian-qian, LIU Qiao-quan, GAO Ji-ping. A Simplified Method for Extracting Genomic DNA from Rice Leaves [J]. Biotechnology Bulletin, 2025, 41(1): 74-84. |
| [7] | MA Xiang-rong, MA Xin, CHEN Yin-xun, LONG Qi-fu, WANG Rong, XING Jiang-wa. Diversity of Culturable Halophilic Bacteria in the Chloride Type Kunteyi Salt Lake in the Qaidam Basin [J]. Biotechnology Bulletin, 2024, 40(7): 285-298. |
| [8] | LIU Rong, TIAN Min-yu, LI Guang-ze, TAN Cheng-fang, RUAN Ying, LIU Chun-lin. Identification and Induced-expression Analysis of REVEILLE Family in Brassica napus L. [J]. Biotechnology Bulletin, 2024, 40(6): 161-171. |
| [9] | YU Li-jun, WANG Qiao-mei, PENG Wen-shu, YAN Liang, YANG Rui-juan. Study on the Microbial Community of Rhizosphere Soil in Ancient Tea Garden and Modern Organic Tea Garden in Jingmai Mountain [J]. Biotechnology Bulletin, 2024, 40(5): 237-247. |
| [10] | LEI Mei-ling, RAO Wen-hua, HU Jin-feng, YUE Qi, WU Zu-jian, FAN Guo-cheng. Bacterial Diversity and Structure in Rhizosphere Soil of Citrus Infested with Huanglongbing [J]. Biotechnology Bulletin, 2024, 40(2): 266-276. |
| [11] | DU Jie, HUANG Xuan-yi, ZHANG Yan, JIANG Qing-chun, YU Zhi-he, WANG Yun, LIU Zhong-yu. Composition of Root-associated Bacteria of Polygonum cuspidatum and Their Relationship with the Bioactive Ingredients [J]. Biotechnology Bulletin, 2024, 40(12): 208-217. |
| [12] | LI Qing, SHI Yu-he, ZHU Jue, LI Xiao-ling, HOU Chao-wen, TONG Qiao-zhen. Genetic Diversity Analysis and DNA Fingerprint Construction of Atractylodes macrocephala Germplasm Resources Based on SCoT Molecular Markers [J]. Biotechnology Bulletin, 2024, 40(11): 142-151. |
| [13] | PI Yi-fei, SONG Xin-hui, WANG Xi-lin, LI Jin-jin, SUN Chang-bin, XU Wei. High-throughput Screening System for Functional R-loop Loci Based on R-loop Targeted Editing Technology [J]. Biotechnology Bulletin, 2024, 40(10): 181-190. |
| [14] | ZHOU Hui-wen, WU Lan-hua, HAN De-peng, ZHENG Wei, YU Pao-lan, WU Yang, XIAO Xiao-jun. Genome-wide Association Study of Seed Glucosinolate Content in Brassica napus [J]. Biotechnology Bulletin, 2024, 40(1): 222-230. |
| [15] | HUANG Xiao-long, SUN Gui-lian, MA Dan-dan, YAN Hui-qing. Construction of Yeast One-hybrid Library and Screening of Factors Regulating LAZY1 Expression in Rice [J]. Biotechnology Bulletin, 2023, 39(9): 126-135. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||