








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (4): 98-105.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0101
Previous Articles Next Articles
HOU Ya-tao1( ), LI Ying-hui1, DENG Lei2, LI Chang-bao3, LI Chuan-you2, SUN Chuan-long4(
), LI Ying-hui1, DENG Lei2, LI Chang-bao3, LI Chuan-you2, SUN Chuan-long4( )
)
Received:2025-01-23
Online:2025-04-26
Published:2025-04-25
Contact:
SUN Chuan-long
E-mail:22220951310005@hainanu.edu.cn;clsun@sdau.edu.cn
HOU Ya-tao, LI Ying-hui, DENG Lei, LI Chang-bao, LI Chuan-you, SUN Chuan-long. Development of Functional Molecular Markers for Tomato Fruit Weight Gene and Population Genotyping Analysis[J]. Biotechnology Bulletin, 2025, 41(4): 98-105.
引物名称 Primer name | 引物序列 Primer sequence (5′‒3′) |
|---|---|
| Fw3.2-pol-F | GTATAAGACTGTTCACTACG |
| Fw3.2-pol-R1 | CGAATCAGCAGTATACACTG |
| Fw3.2-pol-R2 | GTGGATGTGATGCATCAACT |
| Fw11.3-polF | TCACCGTCATCAATCAAAAT |
| Fw11.3-polR1 | CAATTAATTACTATCGAAACGC |
| Fw11.3-polR2 | CATATGCATTTAGTGAGGTTG |
Table 1 Primer sequences for molecular markers of the Fw3.2 and Fw11.3 gene
引物名称 Primer name | 引物序列 Primer sequence (5′‒3′) |
|---|---|
| Fw3.2-pol-F | GTATAAGACTGTTCACTACG |
| Fw3.2-pol-R1 | CGAATCAGCAGTATACACTG |
| Fw3.2-pol-R2 | GTGGATGTGATGCATCAACT |
| Fw11.3-polF | TCACCGTCATCAATCAAAAT |
| Fw11.3-polR1 | CAATTAATTACTATCGAAACGC |
| Fw11.3-polR2 | CATATGCATTTAGTGAGGTTG |
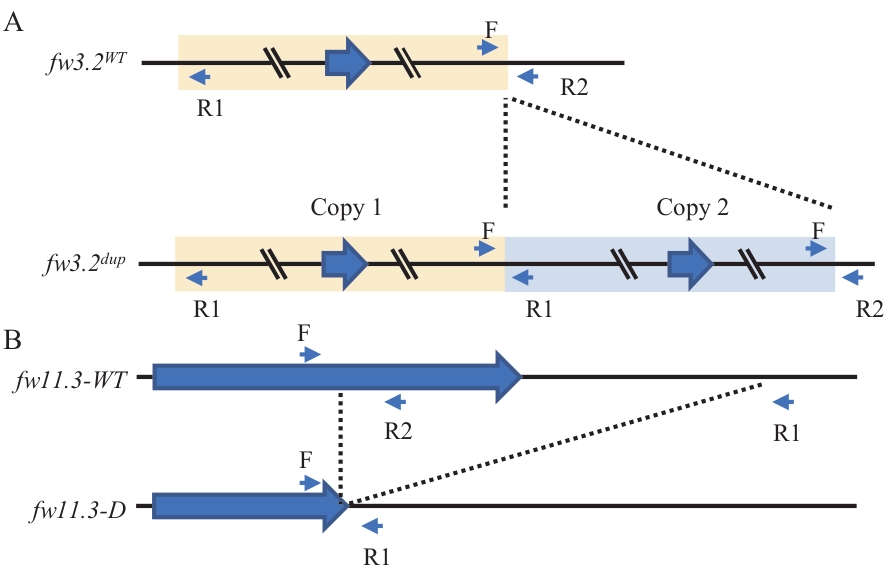
Fig. 1 Schematic diagram of the Fw3.2 (A) and Fw11.3 (B) gene structures and primer locationsfw3.2WT refers to the single-copy allele of the Fw3.2 gene, while fw3.2dup refers to the duplicated allele of the Fw3.2 gene. fw11.3-WT refers to the wild-type allele ofthe Fw11.3 gene, and fw11.3-D indicates the allele with a large fragment deletion in the 3′ region of the Fw11.3 gene. The same below
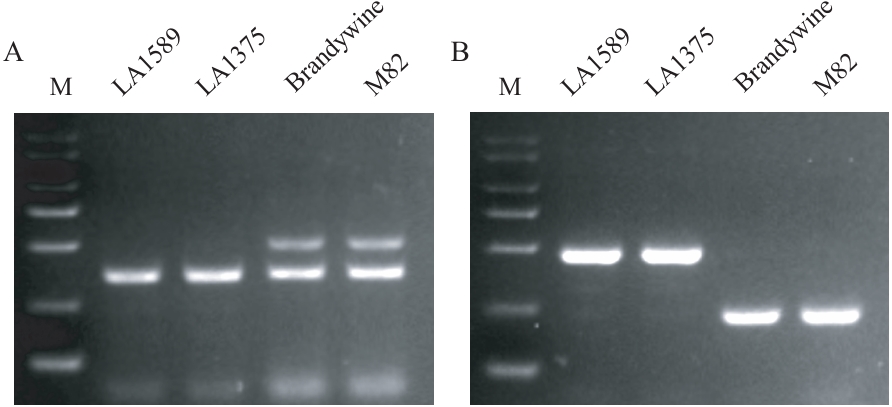
Fig. 2 Polymorphic primer detection for Fw3.2 (A) and Fw11.3 (B) in different tomato materialsM: Marker. LA1589 and LA1375 are S. pimpinellifolium (PIM) accessions, while Brandywine and M82 are big-fruited S. lycopersicum (BIG) accessions
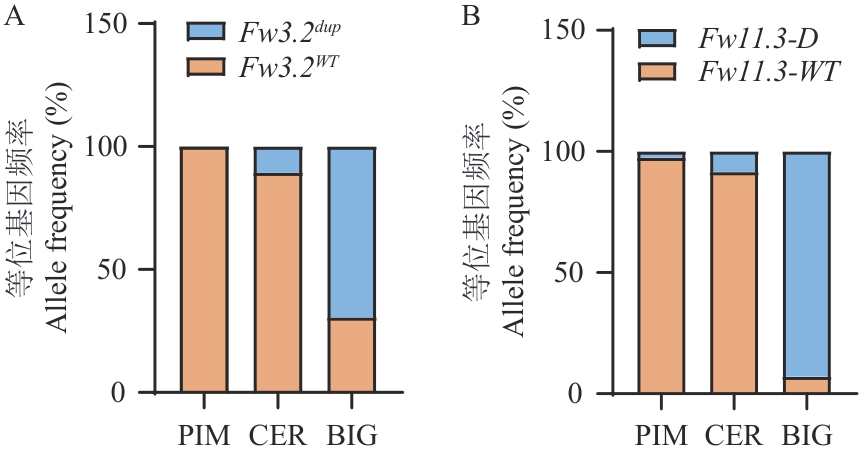
Fig. 3 Allele frequency analysis of Fw3.2 (A) and Fw11.3 (B)A: PIM: S. pimpinellifolium. CER: S. lycopersicum var. cerasiforme. BIG: Big-fruited S. lycopersicum. The same below
地理来源 Geographic origin | 类别 Category | fw3.2WT | fw3.2dup | fw11.3-WT | fw11.3-D |
|---|---|---|---|---|---|
美洲 the Americas | PIM | 76 | 0 | 74 | 2 |
| CER | 83 | 10 | 85 | 8 | |
| BIG | 16 | 3 | 5 | 14 | |
| 亚洲 Asia | BIG | 2 | 48 | 1 | 49 |
Table 2 Genotype analysis of tomato accessions collected from different geographic origins
地理来源 Geographic origin | 类别 Category | fw3.2WT | fw3.2dup | fw11.3-WT | fw11.3-D |
|---|---|---|---|---|---|
美洲 the Americas | PIM | 76 | 0 | 74 | 2 |
| CER | 83 | 10 | 85 | 8 | |
| BIG | 16 | 3 | 5 | 14 | |
| 亚洲 Asia | BIG | 2 | 48 | 1 | 49 |
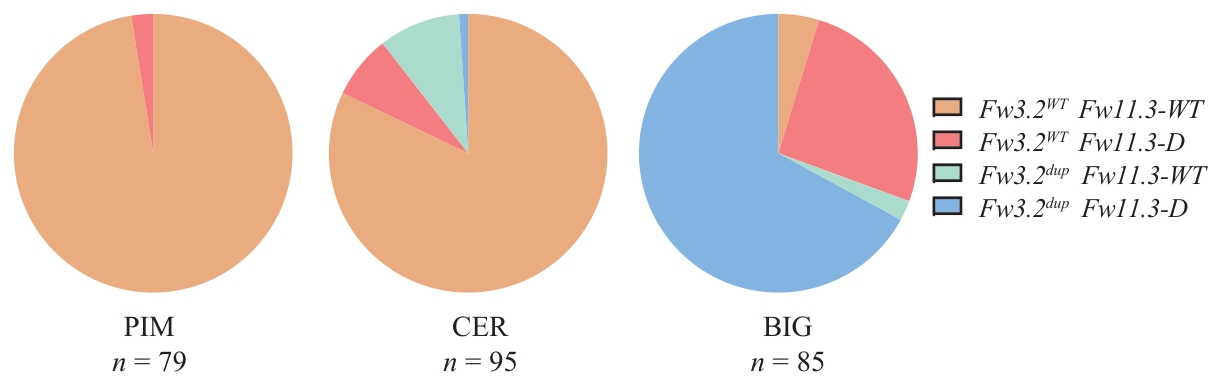
Fig. 4 Haplotype analysis of Fw3.2 and Fw11.3 in different tomato populationsfw3.2WTfw11.3-WT: Haplotype 1; fw3.2WTfw11.3-D: haplotype 2; fw3.2dupfw11.3-WT: haplotype 3; fw3.2dup fw11.3-D: haplotype 4. The same below
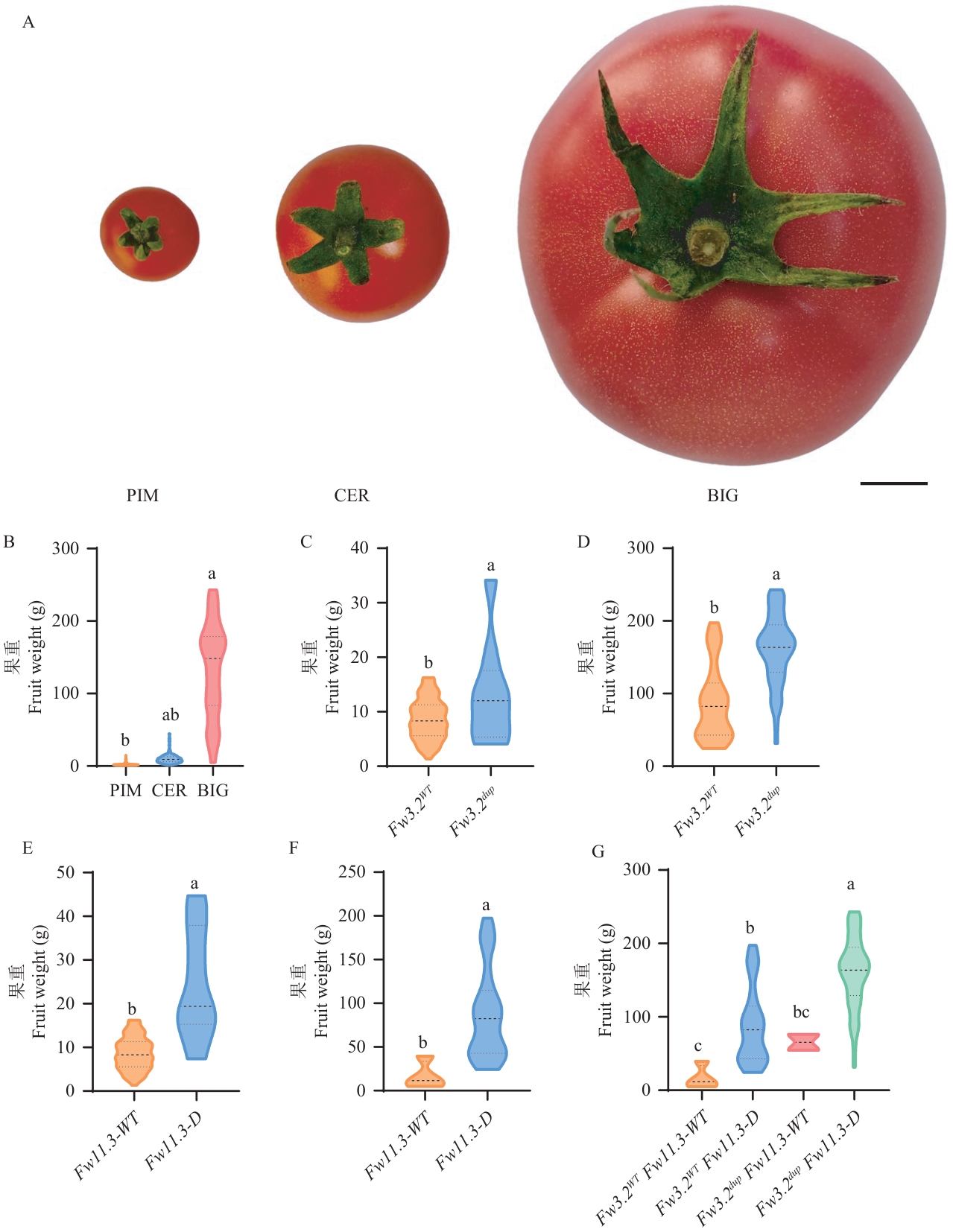
Fig. 5 Phenotypic analysis of fruit weight in different tomato populationsA: Representative images of PIM, CER and BIG varieties. Bar = 1 cm. B: Fruit weight of PIM, CER, and BIG tomatoes. C‒D: Phenotypic variation in fruit weight among distinct Fw3.2 genotypes within CER (C) and BIG (D) populations. E‒F: Phenotypic variation in fruit weight among distinct Fw11.3 genotypes within CER (E) and BIG (F) populations. G: Phenotypic variation in fruit weight among distinct Fw3.2 and Fw11.3 haplotypes within the BIG. Significance of differences was tested using one-way ANOVA for panels B and G, and two-tailed t-tests for panels C‒F. Different letters indicate significant differences (P < 0.05)
| 1 | Chaudhary J, Alisha A, Bhatt V, et al. Mutation breeding in tomato: advances, applicability and challenges [J]. Plants, 2019, 8(5): 128. |
| 2 | 杜敏敏, 周明, 邓磊, 等. 番茄分子育种现状与展望——从基因克隆到品种改良 [J]. 园艺学报, 2017, 44(3): 581-600. |
| Du MM, Zhou M, Deng L, et al. Current status and prospects on tomato molecular breeding—from gene cloning to cultivar improvement [J]. Acta Hortic Sin, 2017, 44(3): 581-600. | |
| 3 | Yang JW, Liu Y, Liang B, et al. Genomic basis of selective breeding from the closest wild relative of large-fruited tomato [J]. Hortic Res, 2023, 10(8): uhad142. |
| 4 | Pereira L, Zhang L, Sapkota M, et al. Unraveling the genetics of tomato fruit weight during crop domestication and diversification [J]. Theor Appl Genet, 2021, 134(10): 3363-3378. |
| 5 | Frary A, Nesbitt TC, Grandillo S, et al. fw2.2: a quantitative trait locus key to the evolution of tomato fruit size [J]. Science, 2000, 289(5476): 85-88. |
| 6 | Beauchet A, Bollier N, Grison M, et al. The cell number regulator FW2.2 protein regulates cell-to-cell communication in tomato by modulating callose deposition at plasmodesmata [J]. Plant Physiol, 2024, 196(2): 883-901. |
| 7 | Cong B, Barrero LS, Tanksley SD. Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication [J]. Nat Genet, 2008, 40(6): 800-804. |
| 8 | Alonge M, Wang XG, Benoit M, et al. Major impacts of widespread structural variation on gene expression and crop improvement in tomato [J]. Cell, 2020, 182(1): 145-161.e23. |
| 9 | Huang ZJ, van der Knaap E. Tomato fruit weight 11.3 maps close to fasciated on the bottom of chromosome 11 [J]. Theor Appl Genet, 2011, 123(3): 465-474. |
| 10 | Mu Q, Huang ZJ, Chakrabarti M, et al. Fruit weight is controlled by cell size regulator encoding a novel protein that is expressed in maturing tomato fruits [J]. PLoS Genet, 2017, 13(8): e1006930. |
| 11 | Rodríguez GR, Muños S, Anderson C, et al. Distribution of SUN, OVATE, LC, and FAS in the tomato germplasm and the relationship to fruit shape diversity [J]. Plant Physiol, 2011, 156(1): 275-285. |
| 12 | Lippman Z, Tanksley SD. Dissecting the genetic pathway to extreme fruit size in tomato using a cross between the small-fruited wild species Lycopersicon pimpinellifolium and L. esculentum var. Giant Heirloom [J]. Genetics, 2001, 158(1): 413-422. |
| 13 | Tanksley SD. The genetic, developmental, and molecular bases of fruit size and shape variation in tomato [J]. Plant Cell, 2004, 16(): S181-S189. |
| 14 | Xu C, Liberatore KL, MacAlister CA, et al. A cascade of arabinosyltransferases controls shoot meristem size in tomato [J]. Nat Genet, 2015, 47(7): 784-792. |
| 15 | Yuste-Lisbona FJ, Fernández-Lozano A, Pineda B, et al. ENO regulates tomato fruit size through the floral meristem development network [J]. Proc Natl Acad Sci USA, 2020, 117(14): 8187-8195. |
| 16 | 孟思达, 韩磊磊, 相恒佐, 等. 番茄心室数的调控机制研究进展 [J]. 园艺学报, 2024, 51(7): 1649-1664. |
| Meng SD, Han LL, Xiang HZ, et al. Research progress on the mechanism of regulating the number of tomato locules [J]. Acta Hortic Sin, 2024, 51(7): 1649-1664. | |
| 17 | Lin T, Zhu GT, Zhang JH, et al. Genomic analyses provide insights into the history of tomato breeding [J]. Nat Genet, 2014, 46(11): 1220-1226. |
| 18 | Razifard H, Ramos A, Della Valle AL, et al. Genomic evidence for complex domestication history of the cultivated tomato in Latin America [J]. Mol Biol Evol, 2020, 37(4): 1118-1132. |
| 19 | Jamann TM, Balint-Kurti PJ, Holland JB. QTL mapping using high-throughput sequencing [J]. Methods Mol Biol, 2015, 1284: 257-285. |
| 20 | Li ZQ, Xu YH. Bulk segregation analysis in the NGS era: a review of its teenage years [J]. Plant J, 2022, 109(6): 1355-1374. |
| 21 | Gao L, Gonda I, Sun HH, et al. The tomato pan-genome uncovers new genes and a rare allele regulating fruit flavor [J]. Nat Genet, 2019, 51(6): 1044-1051. |
| 22 | Li N, He Q, Wang J, et al. Super-pangenome analyses highlight genomic diversity and structural variation across wild and cultivated tomato species [J]. Nat Genet, 2023, 55(5): 852-860. |
| 23 | Consortium TG. The tomato genome sequence provides insights into fleshy fruit evolution [J]. Nature, 2012, 485(7400): 635-641. |
| 24 | Zhou Y, Zhang ZY, Bao ZG, et al. Graph pangenome captures missing heritability and empowers tomato breeding [J]. Nature, 2022, 606(7914): 527-534. |
| 25 | Wang Y, Sun CL, Ye ZB, et al. The genomic route to tomato breeding: Past, present, and future [J]. Plant Physiol, 2024, 195(4): 2500-2514. |
| 26 | Han HY, Li XH, Li TZ, et al. Chromosome-level genome assembly of Solanum pimpinellifolium [J]. Sci Data, 2024, 11(1): 577. |
| 27 | 王娟, 王柏柯, 李宁, 等. 基因编辑技术在番茄育种中的应用进展 [J]. 植物生理学报, 2020, 56(12): 2606-2616. |
| Wang J, Wang BK, Li N, et al. Advances in the application of genome editing in tomato breeding [J]. Plant Physiol J, 2020, 56(12): 2606-2616. | |
| 28 | Zsögön A, Čermák T, Naves ER, et al. De novo domestication of wild tomato using genome editing [J]. Nat Biotechnol, 2018. DOI: 10.1038/nbt.4272 . |
| 29 | Li TD, Yang XP, Yu Y, et al. Domestication of wild tomato is accelerated by genome editing [J]. Nat Biotechnol, 2018. DOI: 10.1038/nbt.4273 . |
| 30 | 刘慧, 黄婷, 陶建平, 等. 番茄生物钟基因SlLNK1、SlEID1和SlELF3光周期响应分析 [J]. 园艺学报, 2023, 50(10): 2079-2090. |
| Liu H, Huang T, Tao JP, et al. Analysis of circadian clock genes SlLNK1, SlEID1, and SlELF3 response to photoperiod in tomato [J]. Acta Hortic Sin, 2023, 50(10): 2079-2090. |
| [1] | LIU Jie, WANG Fei, TAO Ting, ZHANG Yu-jing, CHEN Hao-ting, ZHANG Rui-xing, SHI Yu, ZHANG Yi. Overexpression of SlWRKY41 Improves the Tolerance of Tomato Seedlings to Drought [J]. Biotechnology Bulletin, 2025, 41(2): 107-118. |
| [2] | CHEN Hui-ting, XIA Yi-nan, YAN Wei, ZHANG Meng-fei, CHEN Zhou-mei, HOU Li-xia, LIU Xin. Effect of Exogenous Chlorogenic Acid Treatment on Fruit Quality of Facility Tomato [J]. Biotechnology Bulletin, 2025, 41(2): 119-126. |
| [3] | SONG Ying-pei, WANG Can, ZHOU Hui-wen, KONG Ke-ke, XU Meng-ge, WANG Rui-kai. Analysis of Soybean Pod Dehiscence Habit Based on Whole Genome Association Analysis and Genetic Diversity [J]. Biotechnology Bulletin, 2025, 41(2): 97-106. |
| [4] | LIU Wen-zhi, HE Dan, LI Peng, FU Ying-lin, ZHANG Yi-xin, WEN Hua-jie, YU Wen-qing. Paenibacillus polymyxa New Strain X-11 and Its Growth-promoting Effects on Tomato and Rice [J]. Biotechnology Bulletin, 2024, 40(9): 249-259. |
| [5] | RAO Yong-bin, ZHANG Jun-li. Biological Characteristics, Domestication and Cultivation of Wild Gymnopilus subpurpuratus [J]. Biotechnology Bulletin, 2024, 40(4): 264-270. |
| [6] | ZHAO Yao, WEN Lang, LUO Shao-dan, LI Zi-xing, LIU Chao-chao. Identification of HMA Gene Family and Cadmium Transport Function of SlHMA1 in Tomato [J]. Biotechnology Bulletin, 2024, 40(2): 212-222. |
| [7] | WANG Nan, LIAO Yong-qin, SHI Zhu-feng, SHEN Yun-xin, YANG Tong-yu, FENG Lu-yao, YI Xiao-peng, TANG Jia-cai, CHEN Qi-bin, YANG Pei-wen. Identification of Three Strains of Bacillus from the Forest Soil of Wuliang Mountain and Mining of Their Bioactivities [J]. Biotechnology Bulletin, 2024, 40(2): 277-288. |
| [8] | LI Qing, SHI Yu-he, ZHU Jue, LI Xiao-ling, HOU Chao-wen, TONG Qiao-zhen. Genetic Diversity Analysis and DNA Fingerprint Construction of Atractylodes macrocephala Germplasm Resources Based on SCoT Molecular Markers [J]. Biotechnology Bulletin, 2024, 40(11): 142-151. |
| [9] | SUN Mei-hua, SUN Hui-xian, TIAN Lin-lin, MIAO Yan-xiu, HOU Lei-ping, QI Ming-fang, LI Tian-lai. Functional Identification of YABBY2b Gene and Expression Analysis of Downstream Genes in Tomato [J]. Biotechnology Bulletin, 2024, 40(11): 162-168. |
| [10] | XIAO Liang, WU Zheng-dan, LU Liu-ying, SHI Ping-li, SHANG Xiao-hong, CAO Sheng, ZENG Wen-dan, YAN Hua-bing. Research Progress of Important Traits Genes in Cassava [J]. Biotechnology Bulletin, 2023, 39(6): 31-48. |
| [11] | SHI Jian-lei, ZAI Wen-shan, SU Shi-wen, FU Cun-nian, XIONG Zi-li. Identification and Expression Analysis of miRNA Related to Bacterial Wilt Resistance in Tomato [J]. Biotechnology Bulletin, 2023, 39(5): 233-242. |
| [12] | CHE Yong-mei, GUO Yan-ping, LIU Guang-chao, YE Qing, LI Ya-hua, ZHAO Fang-gui, LIU Xin. Isolation and Identification of Bacterial Strain C8 and B4 and Their Halotolerant Growth-promoting Effects and Mechanisms [J]. Biotechnology Bulletin, 2023, 39(5): 276-285. |
| [13] | HU Ming-yue, YANG Yu, GUO Yang-dong, ZHANG Xi-chun. Functional Analysis of SlMYB96 Gene in Tomato Under Cold Stress [J]. Biotechnology Bulletin, 2023, 39(4): 236-245. |
| [14] | SHEN Yun-xin, SHI Zhu-feng, ZHOU Xu-dong, LI Ming-gang, ZHANG Qing, FENG Lu-yao, CHEN Qi-bin, YANG Pei-wen. Isolation, Identification and Bio-activity of Three Bacillus Strains with Biocontrol Function [J]. Biotechnology Bulletin, 2023, 39(3): 267-277. |
| [15] | XIE Yang, ZHOU Guo-yan, SU Hang, XING Yu-meng, YAN Li-ying. Transcriptome Analysis of Cucumber Seeds Early and Late Germination Under PEG Drought Simulation [J]. Biotechnology Bulletin, 2023, 39(12): 148-157. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||