








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (3): 319-329.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0993
WANG Hao( ), CAO An-ni, GAO Xin-yi, GUO Min-liang(
), CAO An-ni, GAO Xin-yi, GUO Min-liang( )
)
Received:2024-10-12
Online:2025-03-26
Published:2025-03-20
Contact:
GUO Min-liang
E-mail:wanghao@yzu.edu.cn;guoml@yzu.edu.cn
WANG Hao, CAO An-ni, GAO Xin-yi, GUO Min-liang. Enzymatic Characterization and Directed Evolution of Agrobacterium tumefaciens O-demethylase Atu1420[J]. Biotechnology Bulletin, 2025, 41(3): 319-329.

Fig. 1 Identification of Atu1420 expression and catalytic activityA: Schematic diagram of the reaction catalyzed by Atu1420. B: SDS-PAGE electrophoretogram of Atu1420. C: Detecting peak of Atu1420 reaction mixture via HPLC. The blue curve inidicates the reaction mixture without Atu1420, and the red curve indicates the reaction mixture with Atu1420 added. D: Enzyme kinetics curve of Atu1420. The Vmax and Km values are obtained through the double reciprocal plot method
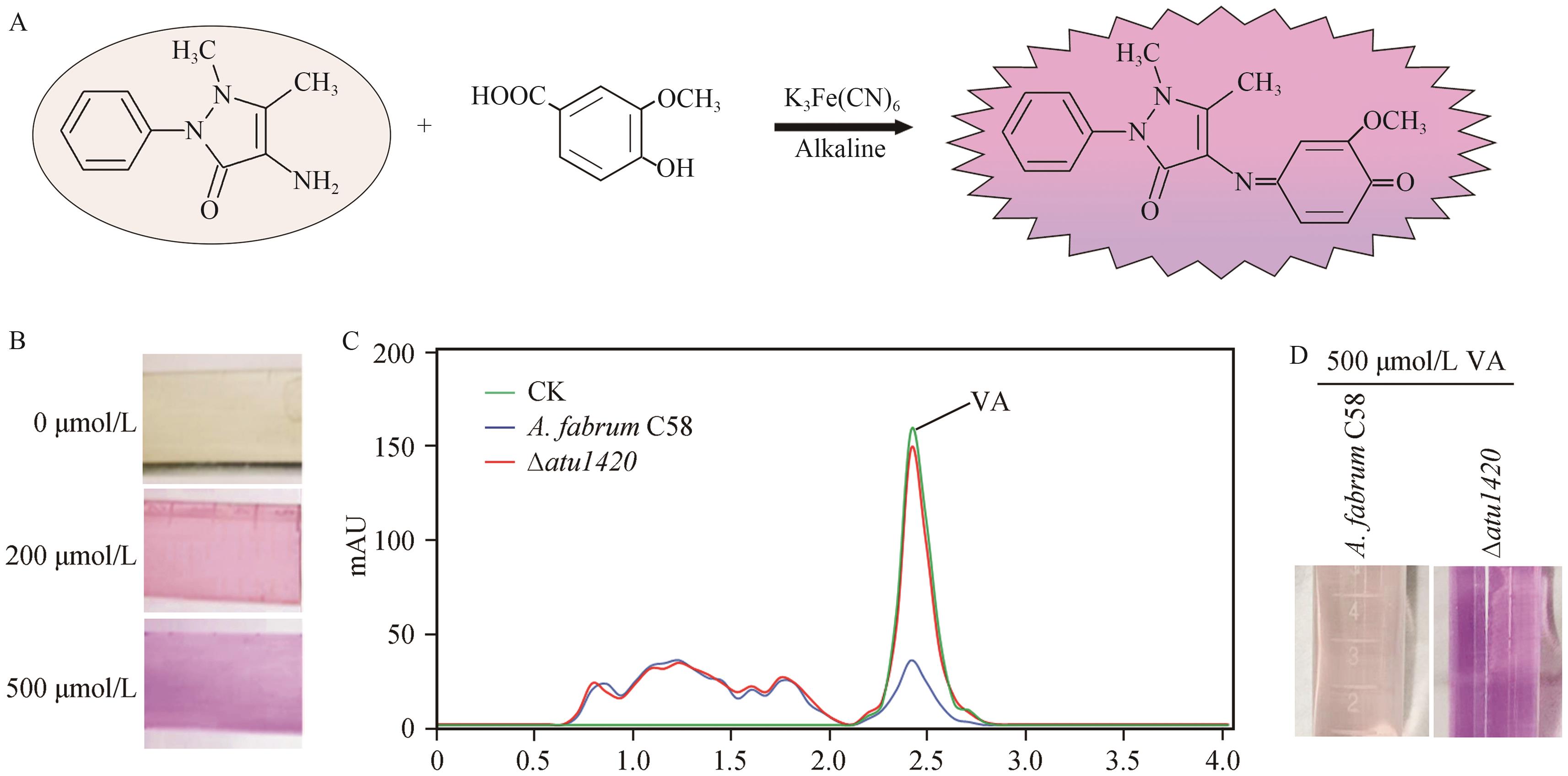
Fig. 3 Establishment of the detection method for VA using 4-aminoantipyrineA: Schematic diagram of the reaction between 4-aminoantipyrine and VA. B: Colors in the reaction between 4-aminoantipyrine and VA of different concentrations. C: HPLC analysis of VA metabolism by A. tumefaciens. CK indicates the untreated control, while the other samples were treated with A. tumefaciens for 6 h. Curves shows the remaining VA levels. D: Detection of VA degradation by A. tumefaciens C58 and ∆atu1420 using the 4-aminoantipyrine method

Fig. 4 Semi-rational design and screening of variants of Atu1420A: Molecular docking of VA with Atu1420. The green molecule refers to Atu1420, with four amino acid residues involved in VA binding and catalysis shown in stick form, where blue indicates nitrogen atoms. The cyan molecule indicates VA, and red indicates oxygen atoms. B: Sequence alignment of Atu1420 homologous proteins. 1 is the Atu1420 homologous protein from Microbacterium trichothecenolyticum, 2 from Sinomonas atrocyanea, 3 from Citricoccus sp. K5, 4 from Sphingobium sp., 5 from Rhizobium leguminosarum, 6 from Betaproteobacteria bacterium, and 7 is Atu1420 itself. Blue triangular arrows indicate the four key amino acid residues from Fig. A, while purple asterisks indicate amino acid residues spatially adjacent to these four key residues. Numbers indicate amino acid positions in Atu1420. C: Catalytic activity of Atu1420 and its variants in the bacteria. CK indicates the control without adding A. tumefaciens, while the other samples were treated with A. tumefaciens for 6 h. D: Catalytic activity of Atu1420 and its variants using proteins. CK indicates the control without adding protein, while the other samples were treated with the protein for 30 min. E: Catalytic activity of multi-points combined variants in the bacteria. CK indicates the control without bacteria, while the other samples were treated with A. tumefaciens for 6 h. F: Catalytic activity of multi-points combined variants using proteins. CK indicates the control without adding protein, while the other samples were treated with the protein for 30 min

Fig. 6 Structural analysis of Atu1420 mutated variantsThe green carbon skeleton indicate the four key amino acid residues of Atu1420, and the carbon skeletons with other colors indicate the four key amino acid residues of other variants. The cyan carbon skeleton molecule indicates VA
| 1 | Mahfuz S, Mun HS, Dilawar MA, et al. Potential role of protocatechuic acid as natural feed additives in farm animal production [J]. Animals, 2022, 12(6): 741. |
| 2 | Kakkar S, Bais S. A review on protocatechuic acid and its pharmacological potential [J]. ISRN Pharmacol, 2014, 2014: 952943. |
| 3 | Masella R, Santangelo C, D'Archivio M, et al. Protocatechuic acid and human disease prevention: biological activities and molecular mechanisms [J]. Curr Med Chem, 2012, 19(18): 2901-2917. |
| 4 | Market and Research 2020. Global Protocatechuic Acid (CAS99-50-3) Market 2020 by Manufacturers, Regions, Type and Application, Forecast to 2025 [R]. Pune: Marketsandresearch.biz, p.156. |
| 5 | Li J, Fu JL, Yue C, et al. Highly efficient biosynthesis of protocatechuic acid via recombinant Pseudomonas putida KT2440 [J]. J Agric Food Chem, 2023, 71(27): 10375-10382. |
| 6 | Quan W, Xu Y, Xie YT, et al. In vitro antioxidant properties and phenolic profile of acid aqueous ethanol extracts from Torreya grandis seed coat [J]. Molecules, 2022, 27(17): 5560. |
| 7 | Antony FM, Wasewar K. Reactive extraction: a promising approach to separate protocatechuic acid [J]. Environ Sci Pollut Res Int, 2020, 27(22): 27345-27357. |
| 8 | 邱笛, 周超, 张根林. 工程微生物合成香草醛的进展与挑战 [J]. 生物加工过程, 2023, 21(4): 355-367. |
| Qiu D, Zhou C, Zhang GL. Advances and challenges in the synthesis of vanillin by engineered microorganisms [J]. Chin J Bioprocess Eng, 2023, 21(4): 355-367. | |
| 9 | 刘超, 邓裕斌, 武书彬. 木质素制备香兰素的方法对比分析 [J]. 造纸科学与技术, 2014, 33(6): 53-57, 99. |
| Liu C, Deng YB, Wu SB. Comparative analysis of vanillin preparation methods from lignin [J]. Pap Sci Technol, 2014, 33(6): 53-57, 99. | |
| 10 | 王鑫鑫, 徐宇骋, 刘翠翠, 等. 一种生物化学法制备原儿茶酸的方法. CN202010921942.9 [P]. 2020-09-04. |
| Wang XX, Xu YC, Liu CC, et al. A method for preparing protocatechuic acid by biochemical method. CN202010921942.9 [P]. 2020-09-04. | |
| 11 | Li K, Frost JW. Synthesis of vanillin from glucose [J]. J Am Chem Soc, 1998, 120(40): 10545-10546. |
| 12 | Wang M, Wang HM, Gao C, et al. Efficient production of protocatechuic acid using systems engineering of Escherichia coli [J]. Metab Eng, 2024, 82: 134-146. |
| 13 | Moriwaki Y, Yato M, Terada T, et al. Understanding the molecular mechanism underlying the high catalytic activity of p-hydroxybenzoate hydroxylase mutants for producing Gallic acid [J]. Biochemistry, 2019, 58(45): 4543-4558. |
| 14 | Upadhyay P, Lali A. Protocatechuic acid production from lignin-associated phenolics [J]. Prep Biochem Biotechnol, 2021, 51(10): 979-984. |
| 15 | Harada A, Kamimura N, Takeuchi K, et al. The crystal structure of a new O-demethylase from Sphingobium sp. strain SYK-6 [J]. FEBS J, 2017, 284(12): 1855-1867. |
| 16 | Rosini E, D'Arrigo P, Pollegioni L. Demethylation of vanillic acid by recombinant LigM in a one-pot cofactor regeneration system [J]. Catal Sci Technol, 2016, 6(21): 7729-7737. |
| 17 | Bleem AC, Kuatsjah E, Johnsen J, et al. Evolution and engineering of pathways for aromatic O-demethylation in Pseudomonas putida KT2440 [J]. Metab Eng, 2024, 84: 145-157. |
| 18 | Bhattacharya A, Sood P, Citovsky V. The roles of plant phenolics in defence and communication during Agrobacterium and Rhizobium infection [J]. Mol Plant Pathol, 2010, 11(5): 705-719. |
| 19 | Lassalle F, Campillo T, Vial L, et al. Genomic species are ecological species as revealed by comparative genomics in Agrobacterium tumefaciens [J]. Genome Biol Evol, 2011, 3: 762-781. |
| 20 | Wang H, Zhang MQ, Wang EY, et al. Agrobacterium fabrum gene atu1420 regulates the pathogenicity by affecting the degradation of growth- and virulence-associated phenols [J]. Res Microbiol, 2023, 174(3): 104011. |
| 21 | Sambrook J, Fristch EF, Manlatis T. Molecular Cloning: A Laboratory Manual [M]. 2nd ed. New York: Cold Spring Harbor Laboratory Press, 1989: 20-25. |
| 22 | Cangelosi GA, Best EA, Martinetti G, et al. Genetic analysis of Agrobacterium [J]. Methods Enzymol, 1991, 204: 384-397. |
| 23 | Gelvin SB. Agrobacterium virulence gene induction [J]. Methods Mol Biol, 2006, 343: 77-84. |
| 24 | Wang H, Zhang MQ, Xu YJ, et al. Agrobacterium fabrum atu0526-encoding protein is the only chemoreceptor that regulates chemoattraction toward the broad antibacterial agent formic acid [J]. Biology, 2021, 10(12): 1345. |
| 25 | Chen H, Huang MF, Yan WL, et al. Enzymatic regio- and enantioselective C-H oxyfunctionalization of fatty acids [J]. ACS Catal, 2021, 11(16): 10625-10630. |
| 26 | Mosae Selvakumar P. Phenol sensing studies by 4-aminoantipyrine method-a review [J]. Org Med Chem Int J, 2018, 5(2): 555657. |
| 27 | Parke D. Supraoperonic clustering of pca genes for catabolism of the phenolic compound protocatechuate in Agrobacterium tumefaciens [J]. J Bacteriol, 1995, 177(13): 3808-3817. |
| 28 | Kohler AC, Mills MJL, Adams PD, et al. Structure of aryl O-demethylase offers molecular insight into a catalytic tyrosine-dependent mechanism [J]. Proc Natl Acad Sci USA, 2017, 114(16): E3205-E3214. |
| 29 | Chica RA, Doucet N, Pelletier JN. Semi-rational approaches to engineering enzyme activity: combining the benefits of directed evolution and rational design [J]. Curr Opin Biotechnol, 2005, 16(4): 378-384. |
| 30 | Senior AW, Evans R, Jumper J, et al. Improved protein structure prediction using potentials from deep learning [J]. Nature, 2020, 577(7792): 706-710. |
| 31 | Reetz M. Making enzymes suitable for organic chemistry by rational protein design [J]. Chembiochem, 2022, 23(14): e202200049. |
| 32 | Savile CK, Janey JM, Mundorff EC, et al. Biocatalytic asymmetric synthesis of chiral amines from ketones applied to sitagliptin manufacture [J]. Science, 2010, 329(5989): 305-309. |
| 33 | Novick SJ, Dellas N, Garcia R, et al. Engineering an amine transaminase for the efficient production of a chiral sacubitril precursor [J]. ACS Catal, 2021, 11(6): 3762-3770. |
| 34 | Xie YY, Xu F, Yang L, et al. Engineering the large pocket of an (S)-selective transaminase for asymmetric synthesis of (S)-1-amino-1-phenylpropane [J]. Catal Sci Technol, 2021, 11(7): 2461-2470. |
| [1] | LU Feng, HUANG Yu-hong, LIN Yan-na, MA Fu-qiang. Advances on Molecular Modifications of Formate Dehydrogenase for CO₂ Reduction [J]. Biotechnology Bulletin, 2025, 41(3): 14-24. |
| [2] | ZHANG Man-yu, DONG Jia-cheng, GOU Fu-fan, GONG Chao-hui, LIU Qian, SUN Wen-liang, KONG zhen, HAO Jie, WANG Min, TIAN Chao-guang. Cloning, Expression, Characterization and Application of the Pectin Esterase MtCE12-1 from Myceliophthora thermophila [J]. Biotechnology Bulletin, 2024, 40(9): 291-300. |
| [3] | ZHENG Fei, YANG Jun-zhao, NIU Yu-feng, LI Rui-lin, ZHAO Guo-zhu. Characterization and Functional Analysis of Lytic Polysaccharide Monooxygenase TtLPMO9I from Thermothelomyces thermophilus [J]. Biotechnology Bulletin, 2024, 40(2): 289-299. |
| [4] | ZHAO Sai-sai, ZHANG Xiao-dan, JIA Xiao-yan, TAO Da-wei, LIU Ke-yu, NING Xi-bin. Investigation on the Complex Mutagenesis Selection of High-yield Nitrate Reductase Strain Staphylococcus simulans ZSJ6 and Its Enzymatic Properties [J]. Biotechnology Bulletin, 2023, 39(4): 103-113. |
| [5] | ZHANG Kai-ping, LIU Yan-li, TU Mian-liang, LI Ji-wei, WU Wen-biao. Optimization of Producing Cellulase by Aspergillus fumigatus A-16 and Its Enzymatic Properties [J]. Biotechnology Bulletin, 2022, 38(9): 215-225. |
| [6] | CHANG Qing, SHU Yue-rong, WANG Wen-tao, JIANG Hao, YAN Quan-de, QIAN Zheng, GAO Xue-chun, WU Jin-hong, ZHANG Yong. Heterologous Expression and Characterization of Endo-type Alginate Lyase from Yeosuana marina sp. JLT21 [J]. Biotechnology Bulletin, 2022, 38(2): 123-131. |
| [7] | TIAN Jia-hui, FENG Jia-li, LU Jun-hua, MAO Lin-jing, HU Zhu-ran, WANG Ying, CHU Jie. Isolation,Purification and Characterization of Laccase LacT-1 from Cerrena unicolor [J]. Biotechnology Bulletin, 2021, 37(8): 186-194. |
| [8] | HAO Jun-yao, MA Fu-qiang, YANG Guang-yu. Functional Analysis of Key Residues in the Active Center of Creatinase from Alcaligenes sp. KS-85 [J]. Biotechnology Bulletin, 2021, 37(3): 75-83. |
| [9] | LIU Shan, YE Wei, ZHU Mu-zi, LI Sai-ni, DENG Zhang-shuang, ZHANG Wei-min. Cloning,Expression and Characterization of a Novel Acyltransferase GPAT [J]. Biotechnology Bulletin, 2021, 37(11): 257-266. |
| [10] | ZHAO Hai-yan, SONG Chen-bin, LIU Zheng-ya, MA Xing-rong, SHANG Hui-hui, LI An-hua, GUAN Xian-jun, WANG Jian-she. Cloning,Recombinant Expression and Enzymatic Properties of α-Amylase Gene from Laceyella sp. [J]. Biotechnology Bulletin, 2020, 36(8): 23-33. |
| [11] | ZHU Cai-lin, LÜ Xiang, XIA Xiao-le. Effect of Site-directed Mutagenesis of Amino Acids in Lid Region on the Enzymatic Properties of T1 Lipase [J]. Biotechnology Bulletin, 2020, 36(11): 94-102. |
| [12] | QIU Jin, HUANG Huo-qing, YAO Bin, LUO Hui-ying. Improvement of Catalytic Activity of Amylase from Bacillus amyloliquefaciens and Its High Expression in Bacillus subtilis [J]. Biotechnology Bulletin, 2019, 35(9): 134-143. |
| [13] | GUO Jing-jing, GUO Lei-lei, ZHAO Yun-xiu, DAI Yi-jun. Research on the NAMase of Ensifer meliloti 1021 and Regulation Mechanism of 3-Cyanopyridine [J]. Biotechnology Bulletin, 2019, 35(8): 51-58. |
| [14] | ZHANG Qing-fang, PANG Fei, YU Shuang, XIAO Jing-hui, DOU Shao-hua, CHI Nai-yu. Screening and Identification of High Uricase-producting Strain from Marine and the Enzymatic Properties [J]. Biotechnology Bulletin, 2019, 35(7): 61-69. |
| [15] | XU Shan ,LI Ren-qiang ,ZHENG Zhen-hua ,ZHANG Yun ,SUN Ai-jun ,HU Yun-feng. Properties of Extracellular Protease of Microbe DH-2 from Mangrove and Optimization of Enzyme Producing Conditions [J]. Biotechnology Bulletin, 2018, 34(6): 120-127. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||