








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (6): 144-154.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0002
ZHANG Yong( ), SONG Sheng-long, LI Yong-tai, ZHANG Xin-yu, LI Yan-jun(
), SONG Sheng-long, LI Yong-tai, ZHANG Xin-yu, LI Yan-jun( )
)
Received:2025-01-02
Online:2025-06-26
Published:2025-06-30
Contact:
LI Yan-jun
E-mail:20222012025@stu.shzu.edu.cn;lyj20022002@sina.com.canalyze
ZHANG Yong, SONG Sheng-long, LI Yong-tai, ZHANG Xin-yu, LI Yan-jun. Cloning of GhSWEET9 in Upland Cotton and Functional Analysis of Resistance to Verticillium Wilt[J]. Biotechnology Bulletin, 2025, 41(6): 144-154.
| 引物 Primer | 序列 Primer sequence (5'-3') | 用途 Application |
|---|---|---|
| CDS-GhSWEET9 | ATGGTTTCGCACCTTGTCAC | 基因克隆 |
CDS-GhSWEET9 Y-SWEET9-F Y-SWEET9-R | AGGTGCTGTGCAGTTCTTT aacacgggggactttgcaacATGGTTTCGCACCTTGTCACCACC tcctcgcccttcacgatacaAGGTGCTGTGCAGTTCTTTTTGGGATC | |
| VIGS-GhSWEET9-F | aaggttaccgaattGAACAGTTCTCCCCAGCC | |
| VIGS-GhSWEET9-R | ctcggtaccggatcAGCTATAATTCCGACCACCATG | |
| PDR-GhSWEET9-F | ccagcctcgagcgg ATGGTTTCGCACCTTGTCAC | |
| PDR-GhSWEET9-R | agctggatccgcgc AGGTGCTGTGCAGTTCTTT | |
| qPCR-GhSWEET9-F | CCATCAATGGCACAGGGACT | 表达量检测 |
| qPCR-GhSWEET9-R | TCGACGTTGAGTGGTGTGAG | |
| UBQ7-F | GAAGGCATTCCACCTGACCAAC | 内参基因扩增 |
| UBQ7-R | CTTGACCTTCTTCTTCTTGTGCTTG |
Table 1 Primers used in this experiment
| 引物 Primer | 序列 Primer sequence (5'-3') | 用途 Application |
|---|---|---|
| CDS-GhSWEET9 | ATGGTTTCGCACCTTGTCAC | 基因克隆 |
CDS-GhSWEET9 Y-SWEET9-F Y-SWEET9-R | AGGTGCTGTGCAGTTCTTT aacacgggggactttgcaacATGGTTTCGCACCTTGTCACCACC tcctcgcccttcacgatacaAGGTGCTGTGCAGTTCTTTTTGGGATC | |
| VIGS-GhSWEET9-F | aaggttaccgaattGAACAGTTCTCCCCAGCC | |
| VIGS-GhSWEET9-R | ctcggtaccggatcAGCTATAATTCCGACCACCATG | |
| PDR-GhSWEET9-F | ccagcctcgagcgg ATGGTTTCGCACCTTGTCAC | |
| PDR-GhSWEET9-R | agctggatccgcgc AGGTGCTGTGCAGTTCTTT | |
| qPCR-GhSWEET9-F | CCATCAATGGCACAGGGACT | 表达量检测 |
| qPCR-GhSWEET9-R | TCGACGTTGAGTGGTGTGAG | |
| UBQ7-F | GAAGGCATTCCACCTGACCAAC | 内参基因扩增 |
| UBQ7-R | CTTGACCTTCTTCTTCTTGTGCTTG |

Fig. 1 Expression pattern of GhSWEET9 gene infected with V. dahliaeCK: Water treatment; Vd991: inoculated with V. dahliae strain Vd991. Significance level (*P<0.05; **P<0.01; ***P<0.001. The same below)
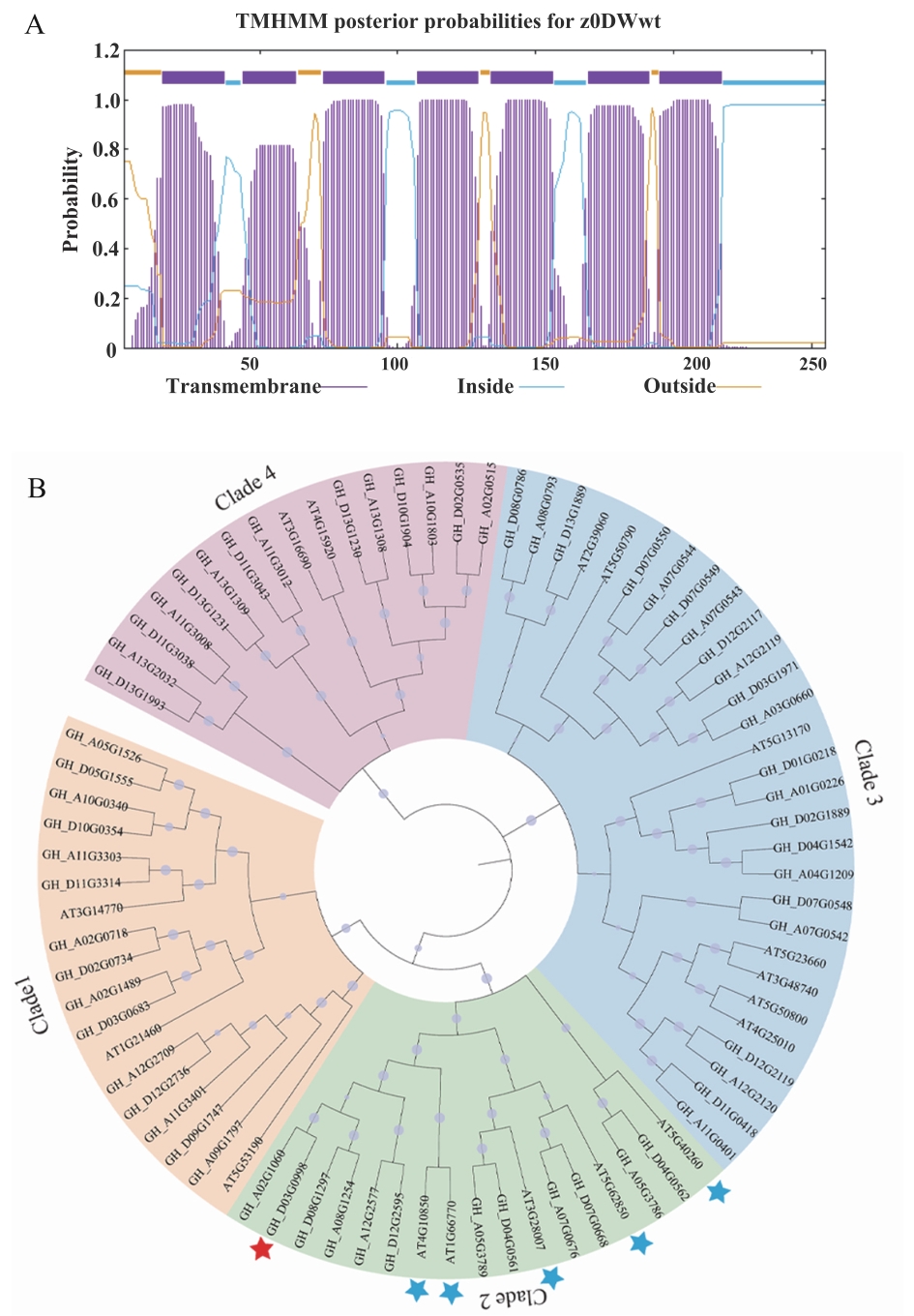
Fig. 2 Bioinformatics analysis of GhSWEET9A: Prediction of the transmembrane domain of GhSWEET9 protein. B: Phylogenetic tree of SWEETs proteins from upland cotton and Arabidopsis
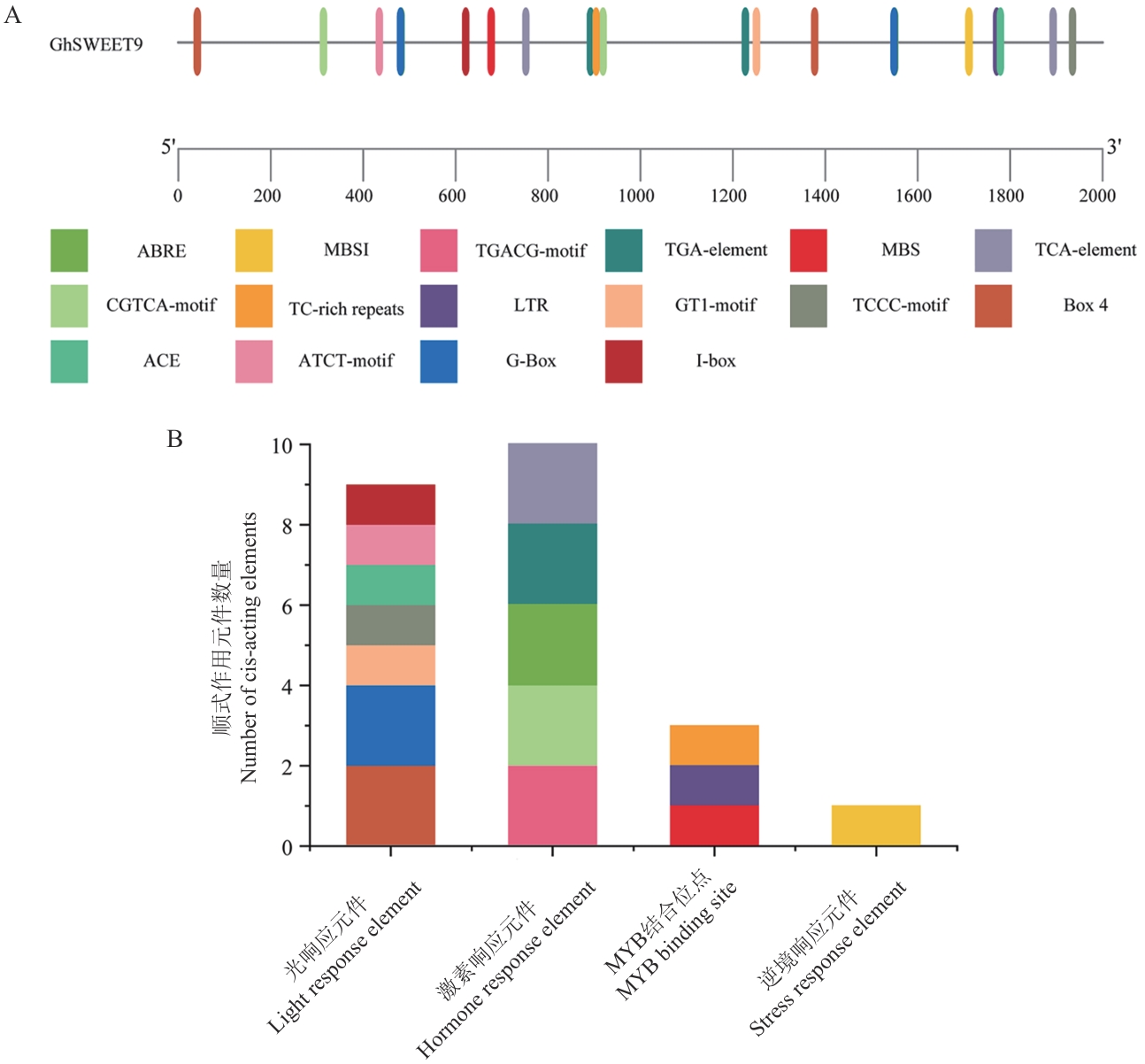
Fig. 3 Analysis of cis-acting elements in the promoter of GhSWEET9 gene in upland cottonA: Analysis of cis-acting elements in the 2 000 bp promoter region upstream of upland cotton GhSWEET9 gene. B: Number statistics of cis-acting elements in different types
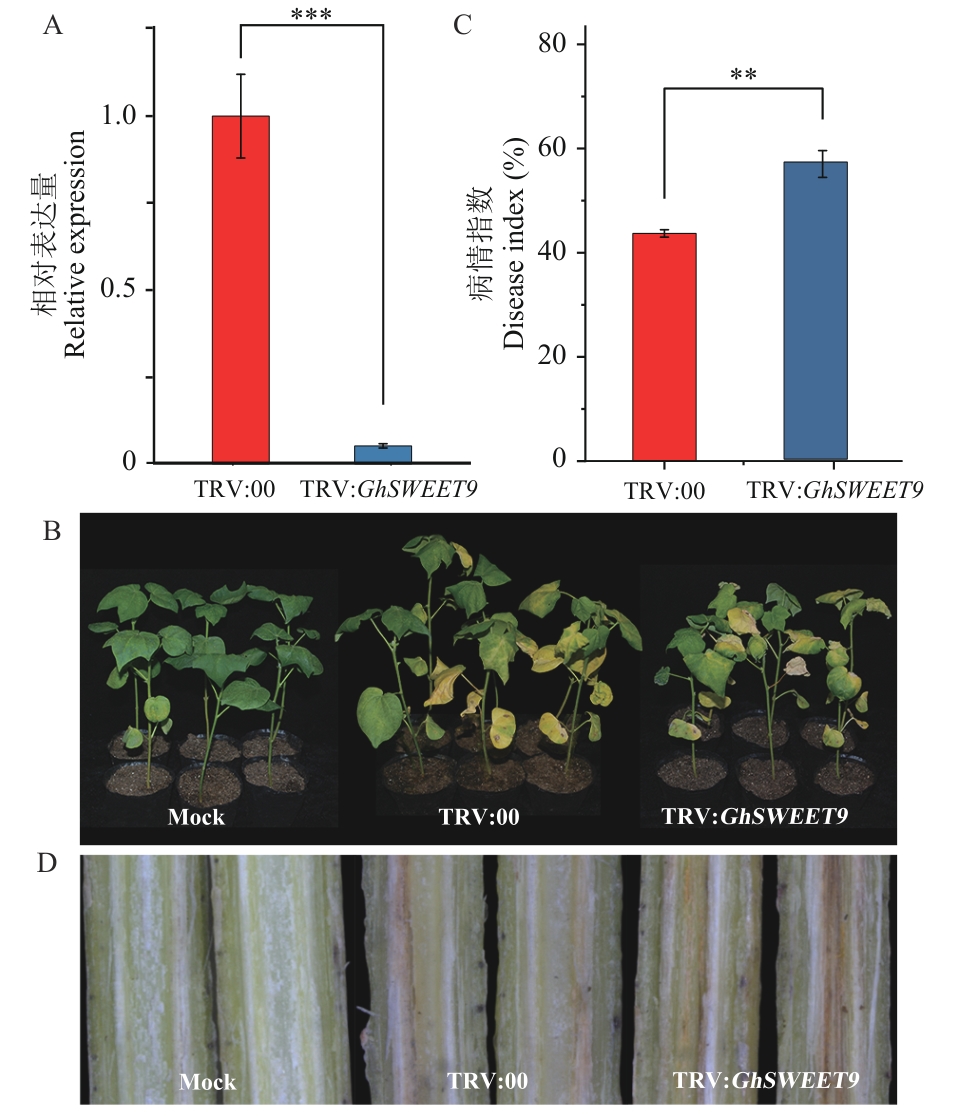
Fig. 6 Identification of GhSWEET9-silenced plants’s resistances to diseaseA: Relative expression of GhSWEET9 in control plants and silenced plants at 14 d post infection. B: Disease symptoms of control plants and silenced plants at 14 d post infection.C: Disease index of control plants and silenced plants at 14 d post infection. D: Vascular bundles browning in stem of control plants and silenced plants at 14 d post infection

Fig. 7 Glucose contents in the roots of control plants and VIGS-silenced plants infected with V. dahliaeA: The glucose content in the roots of cotton plants infected with V. dahliae after 2 d of infection. B: The glucose content in the roots of cotton plants infected with V. dahliae after 6 d of infection. CK: Water treatment; Vd991: inoculated with V. dahliae strain Vd991
| 1 | 刘延财, 唐叶, 吴家和, 等. 生防微生物在棉花黄萎病防治中的研究进展 [J]. 微生物学报, 2025, 65(3): 994-1006. |
| Liu YC, Tan Y, Wu JH, et al. Research progress of biocontrol microbial strains in prevention ofcotton wilt disease [J]. Acta Microbiologica Sinica, 2025, 65(3): 994-1006. | |
| 2 | Zhu YT, Zhao M, Li TT, et al. Interactions between Verticillium dahliae and cotton: pathogenic mechanism and cotton resistance mechanism to Verticillium wilt [J]. Front Plant Sci, 2023, 14: 1174281. |
| 3 | Wilhelm S. Longevity of the Verticillium wilt fungus in the laboratory and field [J]. Phytopathology, 1955, 45(3): 180-181. |
| 4 | 胡丽萍, 张峰, 徐惠, 等. 植物SWEET基因家族结构、功能及调控研究进展 [J]. 生物技术通报, 2017, 33(4): 27-37. |
| Hu LP, Zhang F, Xu H, et al. Research advances in the structure, function and regulation of SWEET gene family in plants [J]. Biotechnol Bull, 2017, 33(4): 27-37. | |
| 5 | Julius BT, Leach KA, Tran TM, et al. Sugar transporters in plants: new insights and discoveries [J]. Plant Cell Physiol, 2017, 58(9): 1442-1460. |
| 6 | Eom JS, Chen LQ, Sosso D, et al. SWEETs, transporters for intracellular and intercellular sugar translocation [J]. Curr Opin Plant Biol, 2015, 25: 53-62. |
| 7 | Chen LQ, Hou BH, Lalonde S, et al. Sugar transporters for intercellular exchange and nutrition of pathogens [J]. Nature, 2010, 468: 527-532. |
| 8 | Chen LQ, Qu XQ, Hou BH, et al. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport [J]. Science, 2012, 335(6065): 207-211. |
| 9 | Yuan M, Wang SP. Rice MtN3/saliva/SWEET family genes and their homologs in cellular organisms [J]. Mol Plant, 2013, 6(3): 665-674. |
| 10 | Zhang XS, Feng CY, Wang MN, et al. Plasma membrane-localized SlSWEET7a and SlSWEET14 regulate sugar transport and storage in tomato fruits [J]. Hortic Res, 2021, 8(1): 186. |
| 11 | Valifard M, Le Hir R, Müller J, et al. Vacuolar fructose transporter SWEET17 is critical for root development and drought tolerance [J]. Plant Physiol, 2021, 187(4): 2716-2730. |
| 12 | Chen LQ, Winnie Lin I, Qu XQ, et al. A cascade of sequentially expressed sucrose transporters in the seed coat and endosperm provides nutrition for the Arabidopsis embryo [J]. Plant Cell, 2015, 27(3): 607-619. |
| 13 | Wang J, Xue XY, Zeng HQ, et al. Sucrose rather than GA transported by AtSWEET13 and AtSWEET14 supports pollen fitness at late anther development stages [J]. New Phytol, 2022, 236(2): 525-537. |
| 14 | Fakher B, Arif Ashraf M, Wang LL, et al. Pineapple SWEET10 is a glucose transporter [J]. Hortic Res, 2023, 10(10): uhad175. |
| 15 | Feng CY, Han JX, Han XX, et al. Genome-wide identification, phylogeny, and expression analysis of the SWEET gene family in tomato [J]. Gene, 2015, 573(2): 261-272. |
| 16 | Gao Y, Wang ZY, Kumar V, et al. Genome-wide identification of the SWEET gene family in wheat [J]. Gene, 2018, 642: 284-292. |
| 17 | Zhao LJ, Yao JB, Chen W, et al. A genome-wide analysis of SWEET gene family in cotton and their expressions under different stresses [J]. J Cotton Res, 2018, 1(1): 7. |
| 18 | Xu ZY, Xu XM, Gong Q, et al. Engineering broad-spectrum bacterial blight resistance by simultaneously disrupting variable TALE-binding elements of multiple susceptibility genes in rice [J]. Mol Plant, 2019, 12(11): 1434-1446. |
| 19 | Liu XZ, Zhang Y, Yang C, et al. AtSWEET4, a hexose facilitator, mediates sugar transport to axial sinks and affects plant development [J]. Sci Rep, 2016, 6: 24563. |
| 20 | Chen HY, Huh JH, Yu YC, et al. The Arabidopsis vacuolar sugar transporter SWEET2 limits carbon sequestration from roots and restricts Pythium infection [J]. Plant J, 2015, 83(6): 1046-1058. |
| 21 | Meteier E, Camera SL, Goddard ML, et al. Overexpression of the VvSWEET4 transporter in grapevine hairy roots increases sugar transport and contents and enhances resistance to Pythium irregulare, a soilborne pathogen [J]. Front Plant Sci, 2019, 10: 884. |
| 22 | Singh J, Das S, Jagadis Gupta K, et al. Physiological implications of SWEETs in plants and their potential applications in improving source-sink relationships for enhanced yield [J]. Plant Biotechnol J, 2023, 21(8): 1528-1541. |
| 23 | Chong JL, Piron MC, Meyer S, et al. The SWEET family of sugar transporters in grapevine: VvSWEET4 is involved in the interaction with Botrytis cinerea [J]. J Exp Bot, 2014, 65(22): 6589-6601. |
| 24 | Sun MX, Zhang ZQ, Ren ZY, et al. The GhSWEET42 glucose transporter participates in Verticillium dahliae infection in cotton [J]. Front Plant Sci, 2021, 12: 690754. |
| 25 | Zhang SL, Dong LJ, Zhang X, et al. The transcription factor GhWRKY70 from Gossypium hirsutum enhances resistance to Verticillium wilt via the jasmonic acid pathway [J]. BMC Plant Biol, 2023, 23(1): 141. |
| 26 | Xu L, Zhang WW, He X, et al. Functional characterization of cotton genes responsive to Verticillium dahliae through bioinformatics and reverse genetics strategies [J]. J Exp Bot, 2014, 65(22): 6679-6692. |
| 27 | Li YT, Li YJ, Yang QW, et al. Dual transcriptome analysis reveals the changes in gene expression in both cotton and Verticillium dahliae during the infection process [J]. J Fungi, 2024, 10(11): 773. |
| 28 | Li W, Ren ZY, Wang ZY, et al. Evolution and stress responses of Gossypium hirsutum SWEET genes [J]. Int J Mol Sci, 2018, 19(3): 769. |
| 29 | Zhang Y, Wang XF, Rong W, et al. Histochemical analyses reveal that stronger intrinsic defenses in Gossypium barbadense than in G. hirsutum are associated with resistance to Verticillium dahliae [J]. Mol Plant Microbe Interact, 2017, 30(12): 984-996. |
| 30 | Gao XQ, Wheeler T, Li ZH, et al. Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt [J]. Plant J, 2011, 66(2): 293-305. |
| 31 | Mo HJ, Wang XF, Zhang Y, et al. Cotton polyamine oxidase is required for spermine and camalexin signalling in the defence response to Verticillium dahliae [J]. Plant J, 2015, 83(6): 962-975. |
| 32 | Breia R, Conde A, Pimentel D, et al. VvSWEET7 is a mono- and disaccharide transporter up-regulated in response to Botrytis cinerea infection in grape berries [J]. Front Plant Sci, 2020, 10: 1753. |
| 33 | Liu QS, Yuan M, Zhou Y, et al. A paralog of the MtN3/saliva family recessively confers race-specific resistance to Xanthomonas oryzae in rice [J]. Plant Cell Environ, 2011, 34(11): 1958-1969. |
| 34 | Cao ZH, Li Z, Meng L, et al. Genome-wide characterization of pyrabactin resistance 1-like (PYL) family genes revealed AhPYL6 confer the resistance to Ralstonia solanacearum in peanut [J]. Plant Physiol Biochem, 2024, 217: 109295. |
| 35 | Xiao W, Zhang Y, Wang Y, et al. The transcription factor TGA2 orchestrates salicylic acid signal to regulate cold-induced proline accumulation in Citrus [J]. Plant Cell, 2024, 37(1): koae290. |
| 36 | Tan ZW, Lu DD, Li L, et al. Comprehensive analysis of safflower R2R3-MYBs reveals the regulation mechanism of CtMYB76 on flavonol biosynthesis [J]. Ind Crops Prod, 2025, 227: 120795. |
| 37 | Liu HB, Liu JX, Si XH, et al. Sugar transporter HmSWEET8 cooperates with HmSTP1 to enhance powdery mildew susceptibility in Heracleum moellendorffii hance [J]. Plants, 2024, 13(16): 2302. |
| 38 | Sosso D, van der Linde K, Bezrutczyk M, et al. Sugar partitioning between Ustilago maydis and its host Zea mays L during infection [J]. Plant Physiol, 2019, 179(4): 1373-1385. |
| 39 | Walerowski P, Gündel A, Yahaya N, et al. Clubroot disease stimulates early steps of phloem differentiation and recruits SWEET sucrose transporters within developing galls [J]. Plant Cell, 2018, 30(12): 3058-3073. |
| 40 | An JY, Zeng T, Ji CY, et al. A Medicago truncatula SWEET transporter implicated in arbuscule maintenance during arbuscular mycorrhizal symbiosis [J]. New Phytol, 2019, 224(1): 396-408. |
| 41 | Chang Q, Liu J, Wang QL, et al. The effect of Puccinia striiformis f.sp. tritici on the levels of water-soluble carbohydrates and the photosynthetic rate in wheat leaves [J]. Physiol Mol Plant Pathol, 2013, 84: 131-137. |
| 42 | Fatima U, Anjali A, Senthil-Kumar M. AtSWEET11 and AtSWEET12: the twin traders of sucrose [J]. Trends Plant Sci, 2022, 27(10): 958-960. |
| 43 | Gao Y, Xue CY, Liu JM, et al. Sheath blight resistance in rice is negatively regulated by WRKY53 via SWEET2a activation [J]. Biochem Biophys Res Commun, 2021, 585: 117-123. |
| 44 | Gao Y, Zhang C, Han X, et al. Inhibition of OsSWEET11 function in mesophyll cells improves resistance of rice to sheath blight disease [J]. Mol Plant Pathol, 2018, 19(9): 2149-2161. |
| 45 | Kim P, Xue CY, Song HD, et al. Tissue-specific activation of DOF11 promotes rice resistance to sheath blight disease and increases grain weight via activation of SWEET14 [J]. Plant Biotechnol J, 2021, 19(3): 409-411. |
| [1] | MA Li-hua, HOU Meng-juan, ZHU Xin-xia. Functional Studies of the Gossypium hirsutumGhNFD4 Gene in Response to Drought in Cotton [J]. Biotechnology Bulletin, 2025, 41(3): 104-111. |
| [2] | DING Ruo-xi, DOU Shuo, AN Ye-zhi, KONG Wen-hui, GUO Wen-jing, ZHANG Dong-mei, WANG Xing-fen, MA Zhi-ying, WU Li-zhu. Extended Research on the Application of VIGS Technology in the Whole Growth Cycle of Cotton [J]. Biotechnology Bulletin, 2025, 41(2): 58-64. |
| [3] | WANG Juan, WANG Xin, TIAN Qin, MA Xiao-mei, ZHOU Xiao-feng, LI Bao-cheng, DONG Cheng-guang. Association Analysis and Exploration of Elite Alleles of Plant Architecture Traits in Gossypium hirsutum L. [J]. Biotechnology Bulletin, 2024, 40(3): 146-154. |
| [4] | HUA Xuan, TIAN Bo-wen, ZHOU Xin-tong, JIANG Zi-han, WANG Shi-qi, HUANG Qian-hui, ZHANG Jian, CHEN Yan-hong. Cloning SmERF B3-45 from Salix matsudana and Functional Analysis on Its Tolerance to Salt [J]. Biotechnology Bulletin, 2024, 40(12): 124-135. |
| [5] | LIU Zhen-yin, DUAN Zhi-zhen, PENG Ting, WANG Tong-xin, WANG Jian. Establishment and Optimization of Virus-induced Gene Silencing System in Bougainvillea peruviana ‘Thimma’ [J]. Biotechnology Bulletin, 2023, 39(7): 123-130. |
| [6] | DONG Cong, GAO Qing-hua, WANG Yue, LUO Tong-yang, WANG Qing-qing. Increasing the Expression of FAD-dependent Glucose Dehydrogenase by Recombinant Pichia pastoris Using a Combined Strategy [J]. Biotechnology Bulletin, 2023, 39(6): 316-324. |
| [7] | LIU Si-jia, WANG Hao-nan, FU Yu-chen, YAN Wen-xin, HU Zeng-hui, LENG Ping-sheng. Cloning and Functional Analysis of LiCMK Gene in Lilium ‘Siberia’ [J]. Biotechnology Bulletin, 2023, 39(3): 196-205. |
| [8] | GAO Kai-yue, GUO Yu-ting, DU Yi-mou, ZHENG Xiao-mei, MA Xin-rong, ZHAO Wei, ZHENG Ping, SUN Ji-bin. A Quantitative Detection Approach for Glucose Uptake in Aspergillus niger: A Case Study of Glucose Transporter MstC [J]. Biotechnology Bulletin, 2023, 39(12): 71-80. |
| [9] | LI Xiu-qing, HU Zi-yao, LEI Jian-feng, DAI Pei-hong, LIU Chao, DENG Jia-hui, LIU Min, SUN Ling, LIU Xiao-dong, LI Yue. Cloning and Functional Analysis of Gene GhTIFY9 Related to Cotton Verticillium Wilt Resistance [J]. Biotechnology Bulletin, 2022, 38(8): 127-134. |
| [10] | NIU Yu-hui, LI Xiang-rong, WU Bei, LI Hong-shan, LI Dian-yu, CHEN Lei, WEI Suo-cheng, FENG Ruo-fei. Effects of Glucose and Sodium Butyrate on the rHSA Yield in CHO-rHSA Engineering Cell Line [J]. Biotechnology Bulletin, 2022, 38(7): 278-286. |
| [11] | LIAO Zhao-min, CAI Jun, LIN Jian-guo, DU Xin, WANG Chang-gao. Expression of Glucose Oxidase Gene from Aspergillus niger in Pichia pastoris and Optimization of Enzyme Production Conditions [J]. Biotechnology Bulletin, 2021, 37(6): 97-107. |
| [12] | XIONG Liang-bin, SUN Ji, LIU Xian-zhou, QU Zhan-guo, JI Yu-qing, XU Yi-xin, WANG Feng-qing. Transcription Variations of Key Genes in the Central Metabolism Pathways of the Sterols Transformation in Mycobacterium [J]. Biotechnology Bulletin, 2021, 37(10): 120-127. |
| [13] | HUO Ming-yue, ZHANG Ge, SU Yu-long, LI Xing, ZHANG Hai-bo, ZHENG Rong, LIU Hao-bao. Isolation of an Acetoin-producing Strain Resistant to High Concentration Glucose and Identification of Metabolites [J]. Biotechnology Bulletin, 2020, 36(8): 53-60. |
| [14] | CHEN Qiao, WU Hai-ying, WANG Zong-shou, XIE Yu-kang, LI Yi-qing, SUN Jun-song. Multiple-site Mutations in Escherichia coli Capable of High-density Growing Induced from the Biosynthesis of Polyhydroxybutyrate [J]. Biotechnology Bulletin, 2020, 36(7): 112-118. |
| [15] | NIU Huan, FENG Jing-yun, HUANG Jian-guo, ZHANG Chao-qun, ZHANG Lu-lu, LIU Xiao-ying, WANG Zhen-ying. Cloning and Functional Analysis of a Wheat Pathogenesis-Related Gene TaSec14 [J]. Biotechnology Bulletin, 2020, 36(6): 54-62. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||