








Biotechnology Bulletin ›› 2026, Vol. 42 ›› Issue (1): 86-94.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0542
Previous Articles Next Articles
YANG Yue-qin1( ), XING Ying1(
), XING Ying1( ), ZHONG Zi-he2,3, TIAN Wei-jun3,4, YANG Xue-qing3,4, WANG Jian-xu3
), ZHONG Zi-he2,3, TIAN Wei-jun3,4, YANG Xue-qing3,4, WANG Jian-xu3
Received:2025-05-26
Online:2026-01-26
Published:2026-02-04
Contact:
XING Ying
E-mail:yangyueqin3569@163.com;xy31034@163.com
YANG Yue-qin, XING Ying, ZHONG Zi-he, TIAN Wei-jun, YANG Xue-qing, WANG Jian-xu. Expression and Functional Analysis of OsMATE34 in Rice under Mercury Stress[J]. Biotechnology Bulletin, 2026, 42(1): 86-94.
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|
| OsMATE34-qF | CATGAGACAAAGCGGCTGTG |
| OsMATE34-qR | TGAACATGGTGGTCACGGAG |
| OsActin-qF | GAGTATGATGAGTCGGGTCCAG |
| OsActin-qR | ACACCAACAATCCCAAACAGAG |
Table 1 Primer sequence for RT-qPCR
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|
| OsMATE34-qF | CATGAGACAAAGCGGCTGTG |
| OsMATE34-qR | TGAACATGGTGGTCACGGAG |
| OsActin-qF | GAGTATGATGAGTCGGGTCCAG |
| OsActin-qR | ACACCAACAATCCCAAACAGAG |
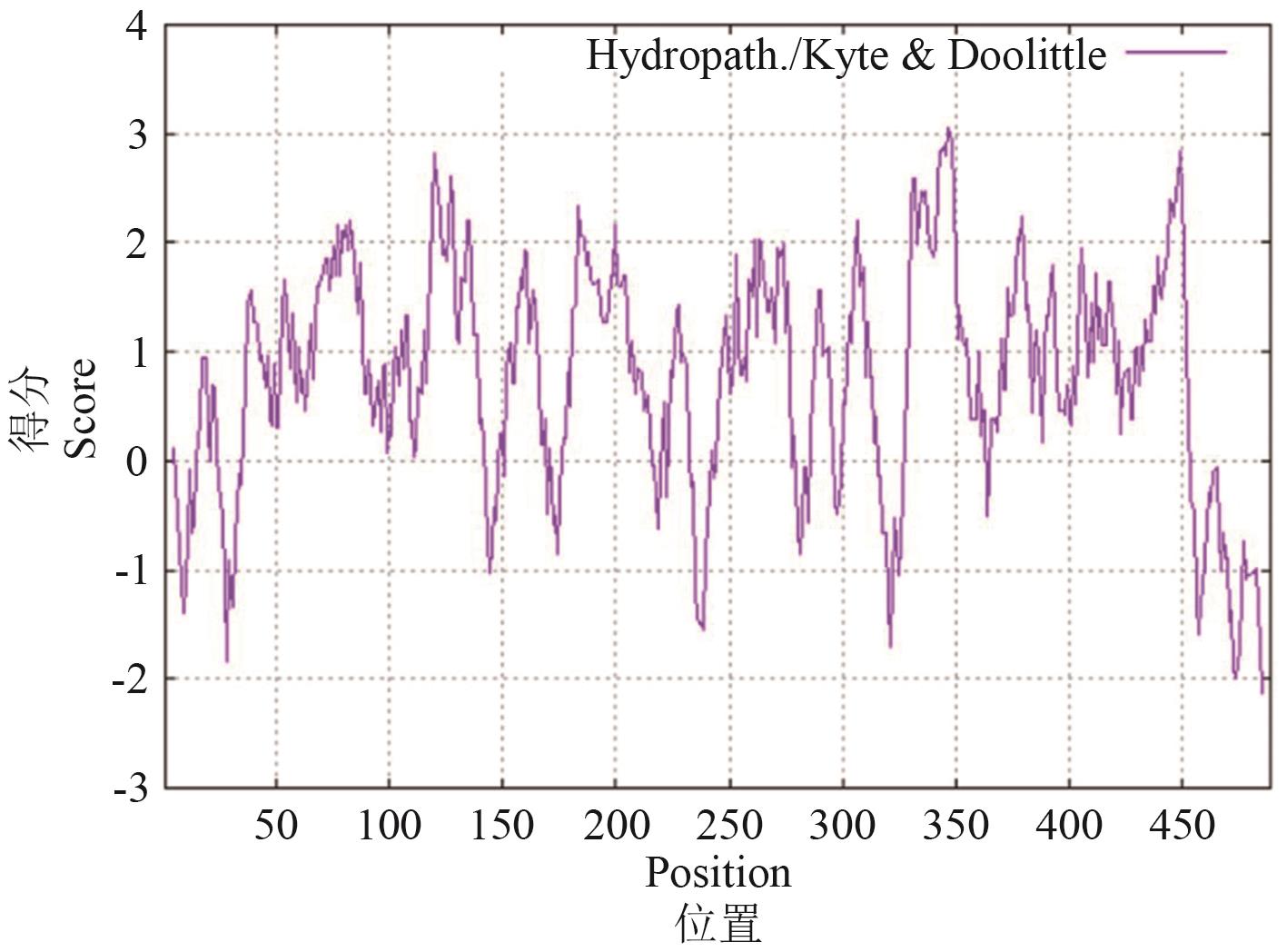
Fig. 1 Analysis of hydrophilicity/hydrophobicity of OsMATE34 proteinRegions with values above 0 are OsMATE34 hydrophobic in character. Regions with values less than 0 are OsMATE34 hydrophilic in character
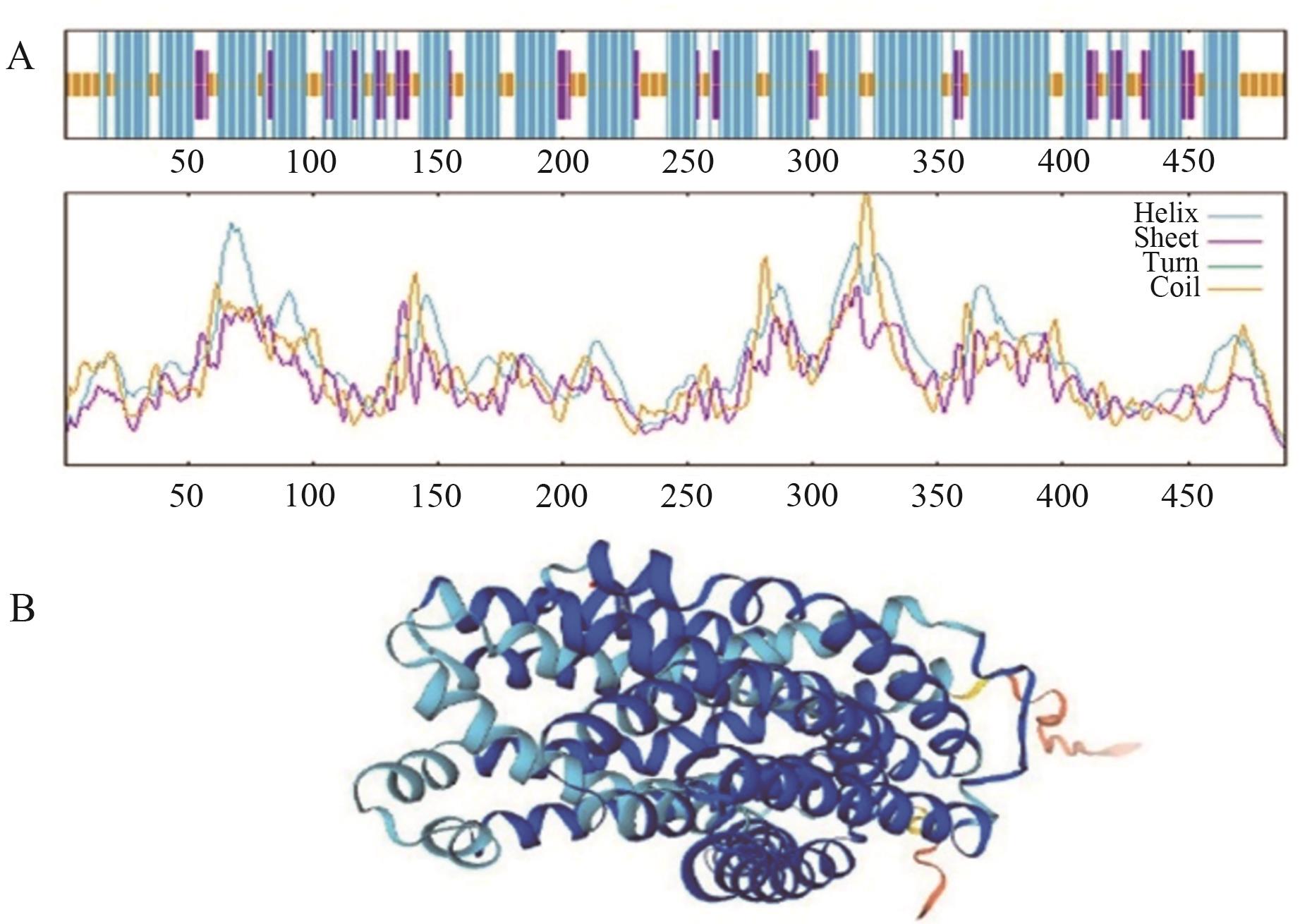
Fig. 2 Secondary and tertiary structure prediction of OsMATE34 proteinA: The secondary structure of OsMATE34, where blue indicates α-helix, purple indicates extended chain, yellow indicates irregular curl, and the abscissa indicates the position of each residue in the amino acid sequence of OsMATE34. B: The tertiary structure of OsMATE34, where dark blue indicates α-helix, light blue indicates irregular curl, and orange indicates extended chain

Fig. 4 Analysis of cis-acting elements of OsMATE34 proteinThe different colored squares indicate a portion of the growth hormone response element (blue), abscisic acid-induced cis-acting element (dark yellow), anaerobic-induced cis-acting element (purple), growth hormone-regulated cis-acting element (pink), methyl jasmonate-induced cis-acting element (green), cis-acting regulator involved in the regulation of zeinolysin metabolism (yellow), cis-acting related to the expression of meristematic tissue action regulatory element (light green), enhancer-like element involved in hypoxia-specific induction (gray), gibberellin-responsive element (light yellow), and MYBHv1 binding site (light pink)
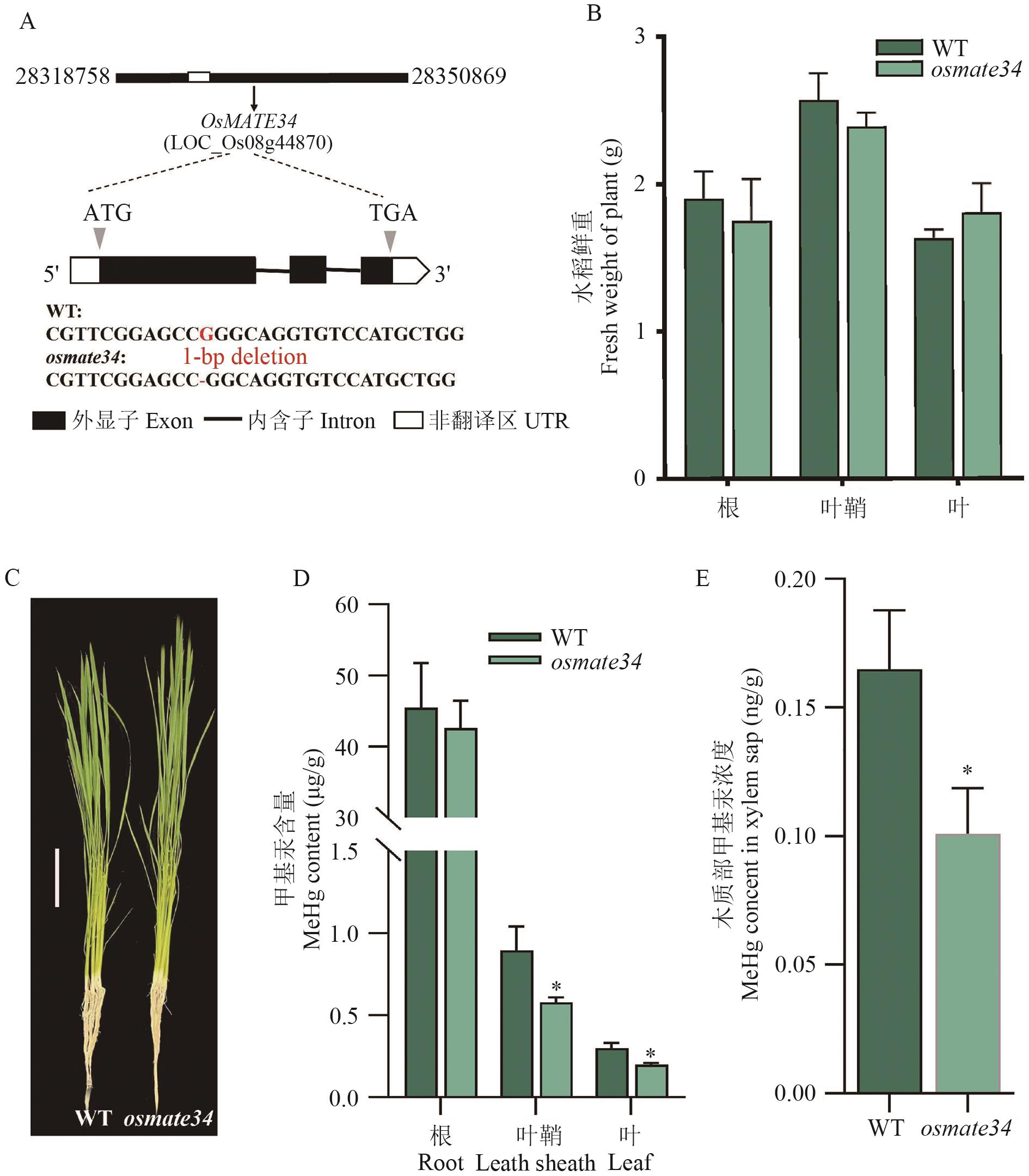
Fig. 8 Growth conditions and MeHg contents of osmate34 and WT under hydroponic conditionsA: Mutated target site of OsMATE34 gene. B: Biomasses of osmate34 and WT in different tissues. C: Plant phenotypes of osmate34 and WT, scale =10 cm. D: MeHg contents of osmate34 and WT in different tissues. E: MeHg contents of osmate34 and WT in xylem sap. * indicates statistically significant difference in comparison to WT at P<0.05
| [1] | Axelrad DA, Bellinger DC, Ryan LM, et al. Dose-response relationship of prenatal mercury exposure and IQ: an integrative analysis of epidemiologic data [J]. Environ Health Perspect, 2007, 115(4): 609-615. |
| [2] | Wang B, Chen M, Ding L, et al. Fish, rice, and human hair mercury concentrations and health risks in typical Hg-contaminated areas and fish-rich areas, China [J]. Environ Int, 2021, 154: 106561. |
| [3] | Beckers F, Rinklebe J. Cycling of mercury in the environment: Sources, fate, and human health implications: a review [J]. Crit Rev Environ Sci Technol, 2017, 47(9): 693-794. |
| [4] | 陆本琦, 刘江, 吕文强, 等. 汞矿区稻田土壤汞形态分布特征及对甲基化的影响 [J]. 矿物岩石地球化学通报, 2021, 40(3): 690-698. |
| Lu BQ, Liu J, Lyu WQ, et al. Distribution characteristics of mercury occurrences in the paddy soil of Hg mining area and its effect on mercury methylation [J]. Bull Mineral Petrol Geochem, 2021, 40(3): 690-698. | |
| [5] | Zhang H, Feng XB, Larssen T, et al. Bioaccumulation of methylmercury versus inorganic mercury in rice (Oryza sativa L.) grain [J]. Environ Sci Technol, 2010, 44(12): 4499-4504. |
| [6] | Feng XB, Li P, Qiu GL, et al. Human exposure to methylmercury through rice intake in mercury mining areas, Guizhou province, China [J]. Environ Sci Technol, 2008, 42(1): 326-332. |
| [7] | Qiu GL, Feng XB, Li P, et al. Methylmercury accumulation in rice (Oryza sativa L.) grown at abandoned mercury mines in Guizhou, China [J]. J Agric Food Chem, 2008, 56(7): 2465-2468. |
| [8] | Li P, Feng XB, Chan HM, et al. Human body burden and dietary methylmercury intake: the relationship in a rice-consuming population [J]. Environ Sci Technol, 2015, 49(16): 9682-9689. |
| [9] | Meng B, Feng XB, Qiu GL, et al. The process of methylmercury accumulation in rice (Oryza sativa L.) [J]. Environ Sci Technol, 2011, 45(7): 2711-2717. |
| [10] | Hao YY, Zhu YJ, Yan RQ, et al. Important roles of thiols in methylmercury uptake and translocation by rice plants [J]. Environ Sci Technol, 2022, 56(10): 6765-6773. |
| [11] | Shoji T. ATP-binding cassette and multidrug and toxic compound extrusion transporters in plants [M]//International Review of Cell and Molecular Biology. Amsterdam: Elsevier, 2014: 303-346. |
| [12] | Huang Y, He GD, Tian WJ, et al. Genome-wide identification of MATE gene family in potato (Solanum tuberosum L.) and expression analysis in heavy metal stress [J]. Front Genet, 2021, 12: 650500. |
| [13] | Morita Y, Kodama K, Shiota S, et al. NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticusand its homolog in Escherichia coli [J]. Antimicrob Agents Chemother, 1998, 42(7): 1778-1782. |
| [14] | Yang H, Guo D, Obianom ON, et al. Multidrug and toxin extrusion proteins mediate cellular transport of cadmium [J]. Toxicol Appl Pharmacol, 2017, 314: 55-62. |
| [15] | Lei GJ, Yokosho K, Yamaji N, et al. Two MATE transporters with different subcellular localization are involved in Al tolerance in buckwheat [J]. Plant Cell Physiol, 2017, 58(12): 2179-2189. |
| [16] | Vasconcellos RCC, Mendes FF, de Oliveira AC, et al. ZmMATE1 improves grain yield and yield stability in maize cultivated on acid soil [J]. Crop Sci, 2021, 61(5): 3497-3506. |
| [17] | Garcia-Oliveira AL, Martins-Lopes P, Tolrá R, et al. Molecular characterization of the citrate transporter gene TaMATE1 and expression analysis of upstream genes involved in organic acid transport under Al stress in bread wheat (Triticum aestivum) [J]. Physiol Plant, 2014, 152(3): 441-452. |
| [18] | Liu JP, Luo XY, Shaff J, et al. A promoter-swap strategy between the AtALMT and AtMATE genes increased Arabidopsis aluminum resistance and improved carbon-use efficiency for aluminum resistance [J]. Plant J, 2012, 71(2): 327-337. |
| [19] | Hoang MTT, Almeida D, Chay S, et al. AtDTX25, a member of the multidrug and toxic compound extrusion family, is a vacuolar ascorbate transporter that controls intracellular iron cycling in Arabidopsis [J]. New Phytol, 2021, 231(5): 1956-1967. |
| [20] | Mackon E, Ma YF, Jeazet Dongho Epse Mackon GC, et al. Computational and transcriptomic analysis unraveled OsMATE34 as a putative anthocyanin transporter in black rice (Oryza sativa L.) caryopsis [J]. Genes, 2021, 12(4): 583. |
| [21] | 王丽, 王万兴, 索海翠, 等. 马铃薯多酚氧化酶基因家族生物信息学及表达分析 [J]. 湖南农业大学学报: 自然科学版, 2019, 45(6): 601-610. |
| Wang L, Wang WX, Suo HC, et al. Bioinformatics and expression analysis of polyphenol oxidase gene family in potato [J]. J Hunan Agric Univ Nat Sci, 2019, 45(6): 601-610. | |
| [22] | Liang L, Horvat M, Cernichiari E, et al. Simple solvent extraction technique for elimination of matrix interferences in the determination of methylmercury in environmental and biological samples by ethylation-gas chromatography-cold vapor atomic fluorescence spectrometry [J]. Talanta, 1996, 43(11): 1883-1888. |
| [23] | Tang ZY, Fan FL, Wang XY, et al. Mercury in rice (Oryza sativa L.) and rice-paddy soils under long-term fertilizer and organic amendment [J]. Ecotoxicol Environ Saf, 2018, 150: 116-122. |
| [24] | 仇广乐, 冯新斌, 梁琏, 等. 溶剂萃取-水相乙基化衍生GC-CVAFS联用测定苔藓样品中的甲基汞 [J]. 分析测试学报, 2005, 24(1): 29-32. |
| Qiu GL, Feng XB, Liang L, et al. Determination of methylmercury in moss by ethylation-gas chromatography-cold vapor atomic fluorescence spectrometry with solvent extraction [J]. J Instrum Anal, 2005, 24(1): 29-32. | |
| [25] | Ikai A. Thermostability and aliphatic index of globular proteins [J]. The Journal of Biochemistry, 1980, 88(6): 1895-1898. |
| [26] | Yazaki K, Sugiyama A, Morita M, et al. Secondary transport as an efficient membrane transport mechanism for plant secondary metabolites [J]. Phytochem Rev, 2008, 7(3): 513-524. |
| [27] | Colmenero-Flores JM, Franco-Navarro JD, Cubero-Font P, et al. Chloride as a beneficial macronutrient in higher plants: new roles and regulation [J]. Int J Mol Sci, 2019, 20(19): 4686. |
| [28] | Inoue SI, Hayashi M, Huang S, et al. A tonoplast-localized magnesium transporter is crucial for stomatal opening in Arabidopsis under high Mg2+ conditions [J]. New Phytol, 2022, 236(3): 864-877. |
| [29] | 方波, 肖腾伟, 苏娜娜, 等. 水稻镉吸收及其在各器官间转运积累的研究进展 [J]. 中国水稻科学, 2021, 35(3): 225-237. |
| Fang B, Xiao TW, Su NN, et al. Research progress on cadmium uptake and its transport and accumulation among organs in rice [J]. Chin J Rice Sci, 2021, 35(3): 225-237. | |
| [30] | 许肖博, 安鹏虎, 郭天骄, 等. 水稻镉胁迫响应机制及防控措施研究进展 [J]. 中国水稻科学, 2021, 35(5): 415-426. |
| Xu XB, An PH, Guo TJ, et al. Research progresses on response mechanisms and control measures of cadmium stress in rice [J]. Chin J Rice Sci, 2021, 35(5): 415-426. | |
| [31] | Li NN, Meng HJ, Xing HT, et al. Genome-wide analysis of MATE transporters and molecular characterization of aluminum resistance in Populus [J]. J Exp Bot, 2017, 68(20): 5669-5683. |
| [32] | Wang JJ, Hou QQ, Li PH, et al. Diverse functions of multidrug and toxin extrusion (MATE) transporters in citric acid efflux and metal homeostasis in Medicago truncatula [J]. Plant J, 2017, 90(1): 79-95. |
| [33] | Li DD, Xu XM, Hu XQ, et al. Genome-wide analysis and heavy metal-induced expression profiling of the HMA gene family in Populus trichocarpa [J]. Front Plant Sci, 2015, 6: 1149. |
| [34] | Strickman RJ, Mitchell CPJ. Accumulation and translocation of methylmercury and inorganic mercury in Oryza sativa: an enriched isotope tracer study [J]. Sci Total Environ, 2017, 574: 1415-1423. |
| [35] | Beauvais-Flück R, Slaveykova VI, Cosio C. Effects of two-hour exposure to environmental and high concentrations of methylmercury on the transcriptome of the macrophyte Elodea nuttallii [J]. Aquat Toxicol, 2018, 194: 103-111. |
| [36] | Kengo Yokosho NY. OsFRDL1 is a citrate transporter required for efficient translocation of iron in rice [J]. Plant Physiol, 2009, 149(1): 297-305. |
| [37] | Das N, Bhattacharya S, Bhattacharyya S, et al. Expression of rice MATE family transporter OsMATE2 modulates arsenic accumulation in tobacco and rice [J]. Plant Mol Biol, 2018, 98(1): 101-120. |
| [38] | 张永福, 莫丽玲, 徐金会, 等. 葡萄耐铝毒基因MATE的克隆与表达分析 [J]. 江西农业大学学报, 2020, 42(5): 906-914. |
| Zhang YF, Mo LL, Xu JH, et al. Cloning and expression analysis of the aluminum-toxicity-tolerance gene MATE in grape [J]. Acta Agric Univ Jiangxiensis, 2020, 42(5): 906-914. | |
| [39] | Yokosho K, Yamaji N, Ma JF. OsFRDL1 expressed in nodes is required for distribution of iron to grains in rice [J]. J Exp Bot, 2016, 67(18): 5485-5494. |
| [1] | REN Yun-er, WU Guo-qiang, CHENG Bin, WEI Ming. Genome-wide Identification of the BvATGs Genes Family in Sugar Beet (Beta vulgaris L.) and Analysis of Their Expression Pattern under Salt Stress [J]. Biotechnology Bulletin, 2026, 42(1): 184-197. |
| [2] | FEI Si-tian, HOU Ying-xiang, LI Lan, ZHANG Chao. Biological Functions and Regulatory Network of SLR1, a Negative Regulator of Gibberellin Signaling in Rice [J]. Biotechnology Bulletin, 2026, 42(1): 13-30. |
| [3] | CHEN Jing-huan, FANG Guo-nan, ZHU Wen-hao, YE Guang-ji, SU Wang, HE Miao-miao, YANG Sheng-long, ZHOU Yun. Starch Characterization and Related Gene Expression Analysis of Potato Germplasm Resources [J]. Biotechnology Bulletin, 2026, 42(1): 170-183. |
| [4] | LI Shan, MA Deng-hui, MA Hong-yi, YAO Wen-kong, YIN Xiao. Identification and Expression Analysis of SKP1 Gene Family in Grapevine (Vitis vinifera L.) [J]. Biotechnology Bulletin, 2025, 41(9): 147-158. |
| [5] | GONG Hui-ling, XING Yu-jie, MA Jun-xian, CAI Xia, FENG Zai-ping. Identification of Laccase (LAC) Gene Family in Potato (Solanum tuberosum L.) and Its Expression Analysis under Salt Stresses [J]. Biotechnology Bulletin, 2025, 41(9): 82-93. |
| [6] | HUA Wen-ping, LIU Fei, HAO Jia-xin, CHEN Chen. Identification and Expression Patterns Analysis of ADH Gene Family in Salvia miltiorrhiza [J]. Biotechnology Bulletin, 2025, 41(8): 211-219. |
| [7] | BAI Yu-guo, LI Wan-di, LIANG Jian-ping, SHI Zhi-yong, LU Geng-long, LIU Hong-jun, NIU Jing-ping. Growth-promoting Mechanism of Trichoderma harzianum T9131 on Astragalus membranaceus Seedlings [J]. Biotechnology Bulletin, 2025, 41(8): 175-185. |
| [8] | REN Rui-bin, SI Er-jing, WAN Guang-you, WANG Jun-cheng, YAO Li-rong, ZHANG Hong, MA Xiao-le, LI Bao-chun, WANG Hua-jun, MENG Ya-xiong. Identification and Expression Analysis of GH17 Gene Family of Pyrenophora graminea [J]. Biotechnology Bulletin, 2025, 41(8): 146-154. |
| [9] | ZENG Dan, HUANG Yuan, WANG Jian, ZHANG Yan, LIU Qing-xia, GU Rong-hui, SUN Qing-wen, CHEN Hong-yu. Genome-wide Identification and Expression Analysis of bZIP Transcription Factor Family in Dendrobium officinale [J]. Biotechnology Bulletin, 2025, 41(8): 197-210. |
| [10] | DENG Mei-bi, YAN Lang, ZHAN Zhi-tian, ZHU Min, HE Yu-bing. Efficient CRISPR Gene Editing in Rice Assisted by RUBY [J]. Biotechnology Bulletin, 2025, 41(8): 65-73. |
| [11] | HOU Ying-xiang, FEI Si-tian, LI Ni, LI Lan, SONG Song-quan, WANG Wei-ping, ZHANG Chao. Research Progress in Response of Rice miRNAs to Biotic Stress [J]. Biotechnology Bulletin, 2025, 41(7): 69-80. |
| [12] | LI Kai-yue, DENG Xiao-xia, YIN Yuan, DU Ya-tong, XU Yuan-jing, WANG Jing-hong, YU Song, LIN Ji-xiang. Identification of LEA Gene Family and Analysis on Its Response to Aluminum Stress in Ricinus communis L. [J]. Biotechnology Bulletin, 2025, 41(7): 128-138. |
| [13] | NIU Jing-ping, ZHAO Jing, GUO Qian, WANG Shu-hong, ZHAO Jin-zhong, DU Wei-jun, YIN Cong-cong, YUE Ai-qin. Identification and Induced Expression Analysis of Transcription Factors NAC in Soybean Resistance to Soybean Mosaic Virus Based on WGCNA [J]. Biotechnology Bulletin, 2025, 41(7): 95-105. |
| [14] | HAN Yi, HOU Chang-lin, TANG Lu, SUN Lu, XIE Xiao-dong, LIANG Chen, CHEN Xiao-qiang. Cloning and Preliminary Functional Analysis of HvERECTA Gene in Hordeum vulgare [J]. Biotechnology Bulletin, 2025, 41(7): 106-116. |
| [15] | GONG Yu-han, CHEN Lan, SHANGFANG Hui-zi, HAO Ling-ying, LIU Shuo-qian. Identification and Expression Profile Analysis of the TRB Gene Family in Tea Plant [J]. Biotechnology Bulletin, 2025, 41(7): 214-225. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||