








Biotechnology Bulletin ›› 2026, Vol. 42 ›› Issue (4): 114-128.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0806
Previous Articles Next Articles
CHEN Deng-ke1,2( ), LAN Gang1, XIA Zhi1, HOU Bao-guo1, YANG Liu-liu1, CAO Cai-rong1, LI Peng-bo1, WU Cui-cui1(
), LAN Gang1, XIA Zhi1, HOU Bao-guo1, YANG Liu-liu1, CAO Cai-rong1, LI Peng-bo1, WU Cui-cui1( )
)
Received:2025-07-26
Online:2026-02-09
Published:2026-02-09
Contact:
WU Cui-cui
E-mail:205436960@qq.com;wucuicui19821021@126.com
CHEN Deng-ke, LAN Gang, XIA Zhi, HOU Bao-guo, YANG Liu-liu, CAO Cai-rong, LI Peng-bo, WU Cui-cui. Identification of ZF-HD Gene Family in Arachis hypogaea and Analysis in Response to Abiotic Stress[J]. Biotechnology Bulletin, 2026, 42(4): 114-128.
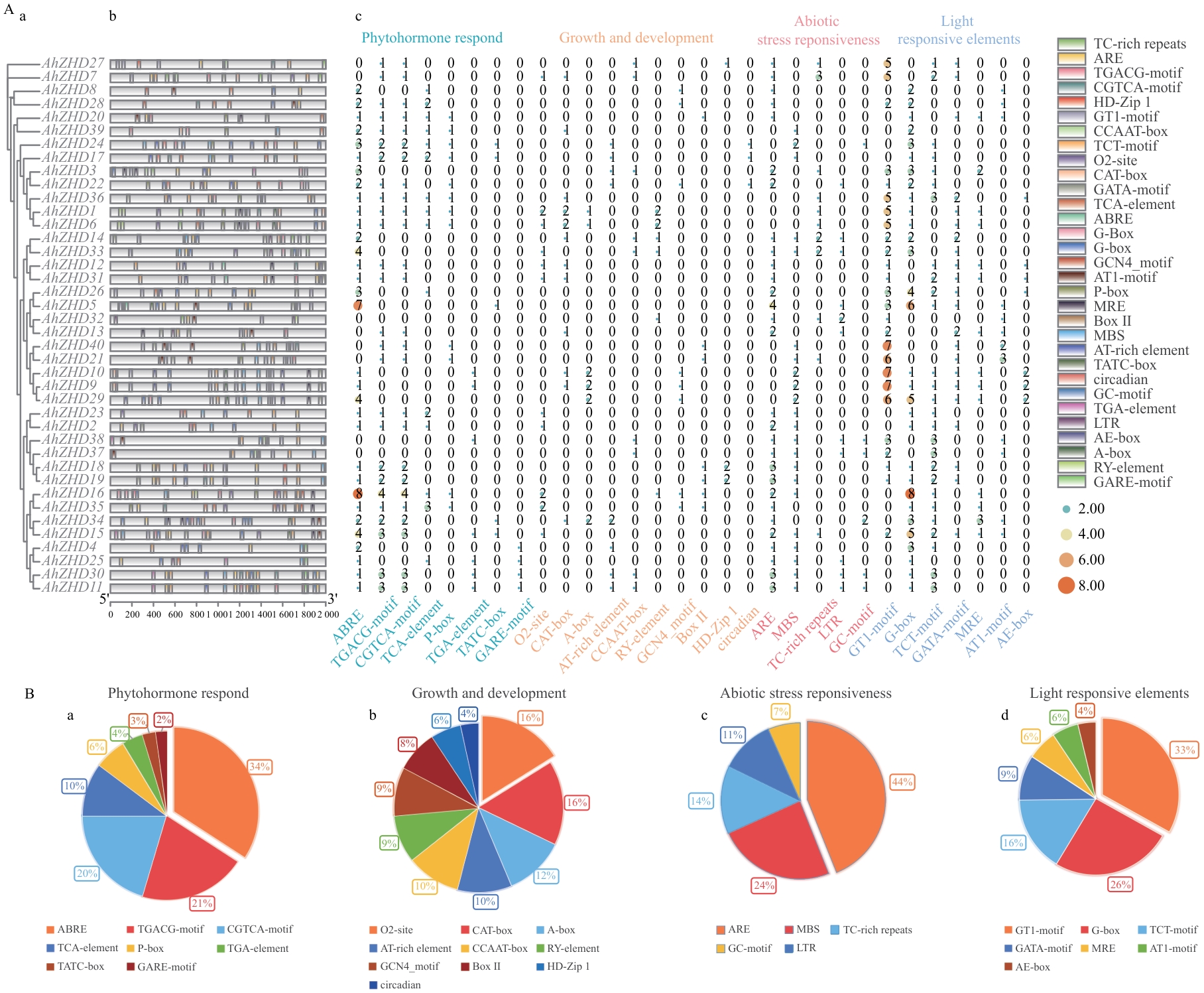
Fig. 3 Analysis of cis-acting elements in the promoter regions of ZF-HD genes in A. hypogaeaA: Cis-acting elements in AhZHD family members (a: Phylogenetic relationship of AhZHD family members. b: Cis-acting elements in the AhZHD family. c: Heatmap of cis-acting elements in the AhZHD family). B: Functional categorization of cis-regulatory elements in the AhZHD family (a: Phytohormone-responsive elements. b: Elements involved in growth and development. c: Abiotic stress-responsive elements. d: Light-responsive elements). The proportion of each specific element type within the respective functional category is indicated as a percentage
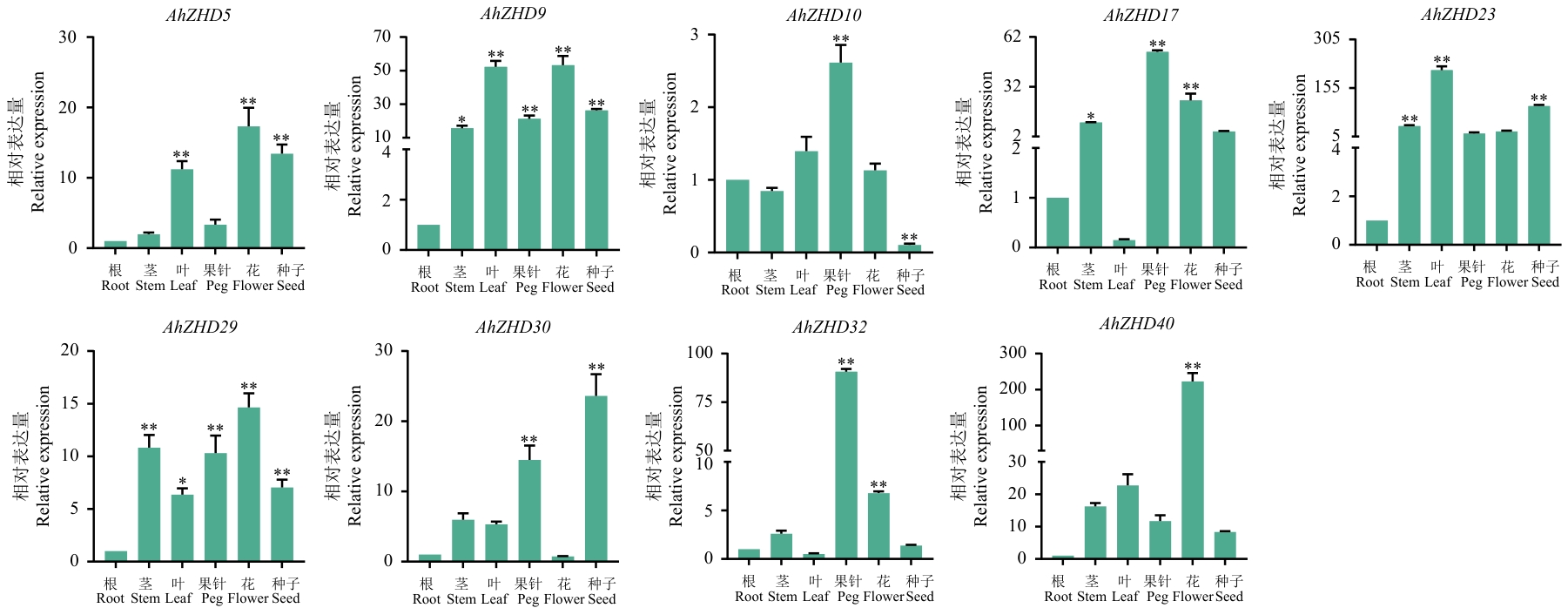
Fig. 10 Expression differences of 9 AhZHDs genes under different tissues and organs based on RT-qPCR* and ** indicate significant differences at the level of 0.05, 0.01 respectively. The same below
| [1] | Wani SH, Anand S, Singh B, et al. WRKY transcription factors and plant defense responses: latest discoveries and future prospects [J]. Plant Cell Rep, 2021, 40(7): 1071-1085. |
| [2] | Dhatterwal P, Sharma N, Prasad M. Decoding the functionality of plant transcription factors [J]. J Exp Bot, 2024, 75(16): 4745-4759. |
| [3] | Thilakarathne AS, Liu F, Zou ZW. Plant signaling hormones and transcription factors: key regulators of plant responses to growth, development, and stress [J]. Plants, 2025, 14(7): 1070. |
| [4] | Piya S, Lopes-Caitar VS, Kim WS, et al. Title: hypermethylation of miRNA genes during nodule development [J]. Front Mol Biosci, 2021, 8: 616623. |
| [5] | Wolberger C. Homeodomain interactions [J]. Curr Opin Struct Biol, 1996, 6(1): 62-68. |
| [6] | Ariel FD, Manavella PA, Dezar CA, et al. The true story of the HD-Zip family [J]. Trends Plant Sci, 2007, 12(9): 419-426. |
| [7] | Hu W, DePamphilis CW, Ma H. Phylogenetic analysis of the plant-specific zinc finger-homeobox and mini zinc finger gene families [J]. J Integr Plant Biol, 2008, 50(8): 1031-1045. |
| [8] | Liu MD, Liu H, Liu WY, et al. Systematic analysis of zinc finger-homeodomain transcription factors (ZF-HDs) in barley (Hordeum vulgare L.) [J]. Genes, 2024, 15(5): 578. |
| [9] | Windhövel A, Hein I, Dabrowa R, et al. Characterization of a novel class of plant homeodomain proteins that bind to the C4 phosphoenolpyruvate carboxylase gene of Flaveria trinervia [J]. Plant Mol Biol, 2001, 45(2): 201-214. |
| [10] | Tan QK, Irish VF. The Arabidopsis zinc finger-homeodomain genes encode proteins with unique biochemical properties that are coordinately expressed during floral development [J]. Plant Physiol, 2006, 140(3): 1095-1108. |
| [11] | Lee YK, Kumari S, Olson A, et al. Role of a ZF-HD transcription factor in miR157-mediated feed-forward regulatory module that determines plant architecture in Arabidopsis [J]. Int J Mol Sci, 2022, 23(15): 8665. |
| [12] | Islam MAU, Nupur JA, Khalid MHB, et al. Genome-wide identification and in silico analysis of ZF-HD transcription factor genes in Zea mays L [J]. Genes, 2022, 13(11): 2112. |
| [13] | Wang WL, Wu P, Li Y, et al. Genome-wide analysis and expression patterns of ZF-HD transcription factors under different developmental tissues and abiotic stresses in Chinese cabbage [J]. Mol Genet Genomics, 2016, 291(3): 1451-1464. |
| [14] | Zhang KP, Yan RQ, Feng XQ, et al. Genome-wide identification and expression analysis of ZF-HD family in sunflower (Helianthus annuus L.) under drought and salt stresses [J]. BMC Plant Biol, 2025, 25(1): 140. |
| [15] | Zhou CZ, Zhu C, Xie SY, et al. Genome-wide analysis of zinc finger motif-associated homeodomain (ZF-HD) family genes and their expression profiles under abiotic stresses and phytohormones stimuli in tea plants (Camellia sinensis) [J]. Sci Hortic, 2021, 281: 109976. |
| [16] | Barth O, Vogt S, Uhlemann R, et al. Stress induced and nuclear localized HIPP26 from Arabidopsis thaliana interacts via its heavy metal associated domain with the drought stress related zinc finger transcription factor ATHB29 [J]. Plant Mol Biol, 2009, 69(1/2): 213-226. |
| [17] | Li QF, Ren YZ, Long JH, et al. Genome-wide identification of zinc finger-homeodomain (ZF-HD) gene family and expression analysis during abiotic stress in rye [J]. Genet Resour Crop Evol, 2025, 72(8): 9927-9943. |
| [18] | Kazerooni EA, Maharachchikumbura SSN, Al-Sadi AM, et al. Effects of the rhizosphere fungus Cunninghamella bertholletiae on the Solanum lycopersicum response to diverse abiotic stresses [J]. Int J Mol Sci, 2022, 23(16): 8909. |
| [19] | Zhao TT, Hu JK, Gao YM, et al. Silencing of the SL-ZH13 transcription factor gene decreases the salt stress tolerance of tomato [J]. J Amer Soc Hort Sci, 2018, 143(5): 391-396. |
| [20] | Liu H, Yang Y, Zhang LS. Zinc finger-homeodomain transcriptional factors (ZF-HDs) in wheat (Triticum aestivum L.): identification, evolution, expression analysis and response to abiotic stresses [J]. Plants, 2021, 10(3): 593. |
| [21] | Lai W, Zhu CX, Hu ZY, et al. Identification and transcriptional analysis of zinc finger-homeodomain (ZF-HD) family genes in cucumber [J]. Biochem Genet, 2021, 59(4): 884-901. |
| [22] | Sun JH, Xie MM, Li XX, et al. Systematic investigations of the ZF-HD gene family in tobacco reveal their multiple roles in abiotic stresses [J]. Agronomy, 2021, 11(3): 406. |
| [23] | 郭曼莉, 李晓彤, 吴澎, 等. 花生加工副产物的综合利用及精深加工 [J]. 粮油食品科技, 2018, 26(3): 27-31. |
| Guo ML, Li XT, Wu P, et al. Comprehensive utilization and deep processing of peanut processing by-products [J]. Sci Technol Cereals Oils Foods, 2018, 26(3): 27-31. | |
| [24] | Sorita GD, Leimann FV, Ferreira SRS. Biorefinery approach: Is it an upgrade opportunity for peanut by-products [J]. Trends Food Sci Technol, 2020, 105: 56-69. |
| [25] | 孙海艳, 侯乾, 董文召, 等. 我国花生品种发展现状 [J]. 中国种业, 2022(7): 15-17. |
| Sun HY, Hou Q, Dong WZ, et al. Development status of peanut varieties in China [J]. China Seed Ind, 2022(7): 15-17. | |
| [26] | 王永慧, 杨久涛, 彭科研, 等. 中国花生产业现状及展望 [J]. 农业展望, 2024, 20(8): 86-91. |
| Wang YH, Yang JT, Peng KY, et al. Current situation and prospects of peanut industry in China [J]. Agric Outlook, 2024, 20(8): 86-91. | |
| [27] | Clevenger J, Chu Y, Scheffler B, et al. A developmental transcriptome map for allotetraploid Arachis hypogaea [J]. Front Plant Sci, 2016, 7: 1446. |
| [28] | Chen CJ, Wu Y, Li JW, et al. TBtools-Ⅱ: a “one for all, all for one” bioinformatics platform for biological big-data mining [J]. Mol Plant, 2023, 16(11): 1733-1742. |
| [29] | Otasek D, Morris JH, Bouças J, et al. Cytoscape Automation: empowering workflow-based network analysis [J]. Genome Biol, 2019, 20(1): 185. |
| [30] | Nicholas Carpita DS. Determination of the pore size of cell walls of living plant cells [J]. Science, 1979, 205(4411): 1144-1147. |
| [31] | Zhang H, Zhao XB, Sun QX, et al. Comparative transcriptome analysis reveals molecular defensive mechanism of Arachis hypogaea in response to salt stress [J]. Int J Genom, 2020, 2020(1): 6524093. |
| [32] | Sparkes IA, Runions J, Kearns A, et al. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants [J]. Nat Protoc, 2006, 1(4): 2019-2025. |
| [33] | Rizwan HM, He JY, Nawaz M, et al. The members of zinc finger-homeodomain (ZF-HD) transcription factors are associated with abiotic stresses in soybean: insights from genomics and expression analysis [J]. BMC Plant Biol, 2025, 25(1): 56. |
| [34] | Jain M, Tyagi AK, Khurana JP. Genome-wide identification, classification, evolutionary expansion and expression analyses of homeobox genes in rice [J]. FEBS J, 2008, 275(11): 2845-2861. |
| [35] | Yang N, Yang ZZ, Zhu ZJ, et al. Identification and expression analysis of ZF-HD transcription factor family and their expression profiles under abiotic and biotic stresses in mango (Mangifera indica) [J]. Physiol Mol Plant Pathol, 2025, 139: 102816. |
| [36] | Xing LX, Peng K, Xue S, et al. Genome-wide analysis of zinc finger-homeodomain (ZF-HD) transcription factors in diploid and tetraploid cotton [J]. Funct Integr Genomics, 2022, 22(6): 1269-1281. |
| [37] | He K, Li CX, Zhang ZY, et al. Genome-wide investigation of the ZF-HD gene family in two varieties of alfalfa (Medicago sativa L.) and its expression pattern under alkaline stress [J]. BMC Genomics, 2022, 23(1): 150. |
| [38] | Bowers JE, Chapman BA, Rong JK, et al. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events [J]. Nature, 2003, 422(6930): 433-438. |
| [39] | Niu HL, Xia PL, Hu YF, et al. Genome-wide identification of ZF-HD gene family in Triticum aestivum: Molecular evolution mechanism and function analysis [J]. PLoS One, 2021, 16(9): e0256579. |
| [40] | 李濯雪, 陈信波. 植物诱导型启动子及相关顺式作用元件研究进展 [J]. 生物技术通报, 2015, 31(10): 8-15. |
| Li ZX, Chen XB. Research advances on plant inducible promoters and related cis-acting elements [J]. Biotechnol Bull, 2015, 31(10): 8-15. | |
| [41] | Ezeh OS, Yamamoto YY. Combinatorial effects of cis-regulatory elements and functions in plants [J]. Rev Agric Sci, 2024, 12: 79-92. |
| [42] | Xu CZ, Luo F, Hochholdinger F. LOB domain proteins: beyond lateral organ boundaries [J]. Trends Plant Sci, 2016, 21(2): 159-167. |
| [43] | Kim MJ, Kim M, Kim J. Combinatorial interactions between LBD10 and LBD27 are essential for male gametophyte development in Arabidopsis [J]. Plant Signal Behav, 2015, 10(8): e1044193. |
| [44] | Zimmermann P, Hirsch-Hoffmann M, Hennig L, et al. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox [J]. Plant Physiol, 2004, 136(1): 2621-2632. |
| [45] | Maffucci T, Falasca M. Specificity in pleckstrin homology (PH) domain membrane targeting: a role for a phosphoinositide-protein co-operative mechanism [J]. FEBS Lett, 2001, 506(3): 173-179. |
| [46] | Yamagishi K, Nagata N, Yee KM, et al. TANMEI/EMB2757 encodes a WD repeat protein required for embryo development in Arabidopsis [J]. Plant Physiol, 2005, 139(1): 163-173. |
| [47] | 游晓慧, 李威, 陶启长, 等. WD40重复蛋白家族基因At1g65030调控拟南芥种子的重量与体积 [J]. 植物生理学报, 2011, 47(7): 715-725. |
| You XH, Li W, Tao QC, et al. At1g65030, a WD40-repeat protein gene, regulates seed mass and size in Arabidopsis [J]. Plant Physiol J, 2011, 47(7): 715-725. | |
| [48] | 舒英杰. 大豆Homeobox基因GmSbh1的分离、表达分析及其与种子劣变的关系 [D]. 南京: 南京农业大学, 2014. |
| Shu YJ. Isolation and expression analysis of soybean Homeobox gene GmSbh1 and its relationship with seed deterioration [D]. Nanjing: Nanjing Agricultural University, 2014. | |
| [49] | Xu XR, Zhou H, Yang QH, et al. ZF-HD gene family in rapeseed (Brassica napus L.): genome-wide identification, phylogeny, evolutionary expansion and expression analyses [J]. BMC Genomics, 2024, 25(1): 1181. |
| [50] | Shi BW, Haq IU, Fiaz S, et al. Genome-wide identification and expression analysis of the ZF-HD gene family in pea (Pisum sativum L.) [J]. Front Genet, 2022, 13: 1089375. |
| [1] | SU Yan-zhu, LI Da, ZHANG Ai-ai, LIU Yong-guang, ZHANG Xiu-rong, XUE Qi-qin. Identification and Expression Analysis of CAD Gene Family in Soybean(Glycine max (L.) Merr.) [J]. Biotechnology Bulletin, 2026, 42(4): 101-113. |
| [2] | YIN Ya-long, ZHANG Ming-yang, WANG Jie-min, MIAO Xue-xue, CHEN Jin, WANG Wei-ping. Advances in Coordinated Tolerance Mechanisms to Abiotic Stresses in Rice [J]. Biotechnology Bulletin, 2026, 42(4): 26-37. |
| [3] | LIU Qing-yuan, WU Hong-qi, CHEN Xiu-e, CHEN Jian, JIANG Yuan-ze, HE Yan-zi, YU Qi-wei, LIU Ren-xiang. Function of Transcription Factor NtMYB96a in Regulating the Tolerance of Tobacco to Drought [J]. Biotechnology Bulletin, 2026, 42(4): 239-250. |
| [4] | NONG Wei-you, ZHAO Chang-zu, QIAN Zhen-feng, DING Qian, WANG Yu-jie, CHEN Shu-ying, HE Li-lian, LI Fu-sheng. Identification of the EfBBX Gene Family in Erianthus fulvus and Analysis of Its Expression Patterns Under Cold Stress [J]. Biotechnology Bulletin, 2026, 42(2): 1-11. |
| [5] | REN Yun-er, WU Guo-qiang, CHENG Bin, WEI Ming. Genome-wide Identification of the BvATGs Genes Family in Sugar Beet (Beta vulgaris L.) and Analysis of Their Expression Pattern under Salt Stress [J]. Biotechnology Bulletin, 2026, 42(1): 184-197. |
| [6] | YANG Yue-qin, XING Ying, ZHONG Zi-he, TIAN Wei-jun, YANG Xue-qing, WANG Jian-xu. Expression and Functional Analysis of OsMATE34 in Rice under Mercury Stress [J]. Biotechnology Bulletin, 2026, 42(1): 86-94. |
| [7] | YANG Juan, FENG Hui, JI Nai-zhe, SUN Li-ping, WANG Yun, ZHANG Jia-nan, ZHAO Shi-wei. Cloning and Functional Analysis of AP2/ERF Transcription Factors RcERF4 and RcRAP2-12 in Rose [J]. Biotechnology Bulletin, 2026, 42(1): 150-160. |
| [8] | FEI Si-tian, HOU Ying-xiang, LI Lan, ZHANG Chao. Biological Functions and Regulatory Network of SLR1, a Negative Regulator of Gibberellin Signaling in Rice [J]. Biotechnology Bulletin, 2026, 42(1): 13-30. |
| [9] | LI Jian-bin, HOU Jia-e, LI Lei-lin, AI Ming-tao, LIU Tian-tai, CUI Xiu-ming, YANG Qian. Genome-wide Identification of Panax notoginseng Lipoxygenases Coupled in Response to Methyl-jasmonate and Wounding [J]. Biotechnology Bulletin, 2026, 42(1): 218-229. |
| [10] | WU Cui-cui, CHEN Deng-ke, LAN Gang, XIA Zhi, LI Peng-bo. Bioinformatics Analysis of Peanut Transcription Factor AhHDZ70 and Its Tolerances to Salt and Drought [J]. Biotechnology Bulletin, 2026, 42(1): 198-207. |
| [11] | CHEN Jing-huan, FANG Guo-nan, ZHU Wen-hao, YE Guang-ji, SU Wang, HE Miao-miao, YANG Sheng-long, ZHOU Yun. Starch Characterization and Related Gene Expression Analysis of Potato Germplasm Resources [J]. Biotechnology Bulletin, 2026, 42(1): 170-183. |
| [12] | LONG Lin-xi, ZENG Yin-ping, WANG Qian, DENG Yu-ping, GE Min-qian, CHEN Yan-zhuo, LI Xin-juan, YANG Jun, ZOU Jian. Identification of Sunflower GH3 Gene Family and Analysis of Their Function in Flower Development [J]. Biotechnology Bulletin, 2026, 42(1): 125-138. |
| [13] | ZENG Ting, ZHANG Lan, LUO Rui. Functional Analysis of the Transcription Factor MpR2R3-MYB17 in Regulating Gemma Development in Marchantia polymorpha L. [J]. Biotechnology Bulletin, 2026, 42(1): 208-217. |
| [14] | HE Qi-chen, YANG Yang, ALIYA Waili, TANG Xin-yue, LI Zhong-xi, CHEN Yong-kun, CHEN Ling-na. Investigation into the Family Characteristics of the Lavender Copper Amine Oxidase Gene and the Role of LaCuAO1 in Bioamine Degradation [J]. Biotechnology Bulletin, 2026, 42(1): 114-124. |
| [15] | LYU Cheng-cong, HENG Meng, CHEN Si-qi, JIN Xue-hua. Cloning and Functional Analysis of ZhGSTF Related to Anthocyanin Transport Zantedeschia hybrida [J]. Biotechnology Bulletin, 2026, 42(1): 161-169. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||