








Biotechnology Bulletin ›› 2026, Vol. 42 ›› Issue (4): 239-250.doi: 10.13560/j.cnki.biotech.bull.1985.2025-1094
LIU Qing-yuan1( ), WU Hong-qi1, CHEN Xiu-e2, CHEN Jian2, JIANG Yuan-ze2, HE Yan-zi1, YU Qi-wei2(
), WU Hong-qi1, CHEN Xiu-e2, CHEN Jian2, JIANG Yuan-ze2, HE Yan-zi1, YU Qi-wei2( ), LIU Ren-xiang1(
), LIU Ren-xiang1( )
)
Received:2025-10-16
Online:2026-02-09
Published:2026-02-09
Contact:
YU Qi-wei, LIU Ren-xiang
E-mail:18188242208@163.com;ycs327@126.com;rxliu@gzu.edu.cn
LIU Qing-yuan, WU Hong-qi, CHEN Xiu-e, CHEN Jian, JIANG Yuan-ze, HE Yan-zi, YU Qi-wei, LIU Ren-xiang. Function of Transcription Factor NtMYB96a in Regulating the Tolerance of Tobacco to Drought[J]. Biotechnology Bulletin, 2026, 42(4): 239-250.
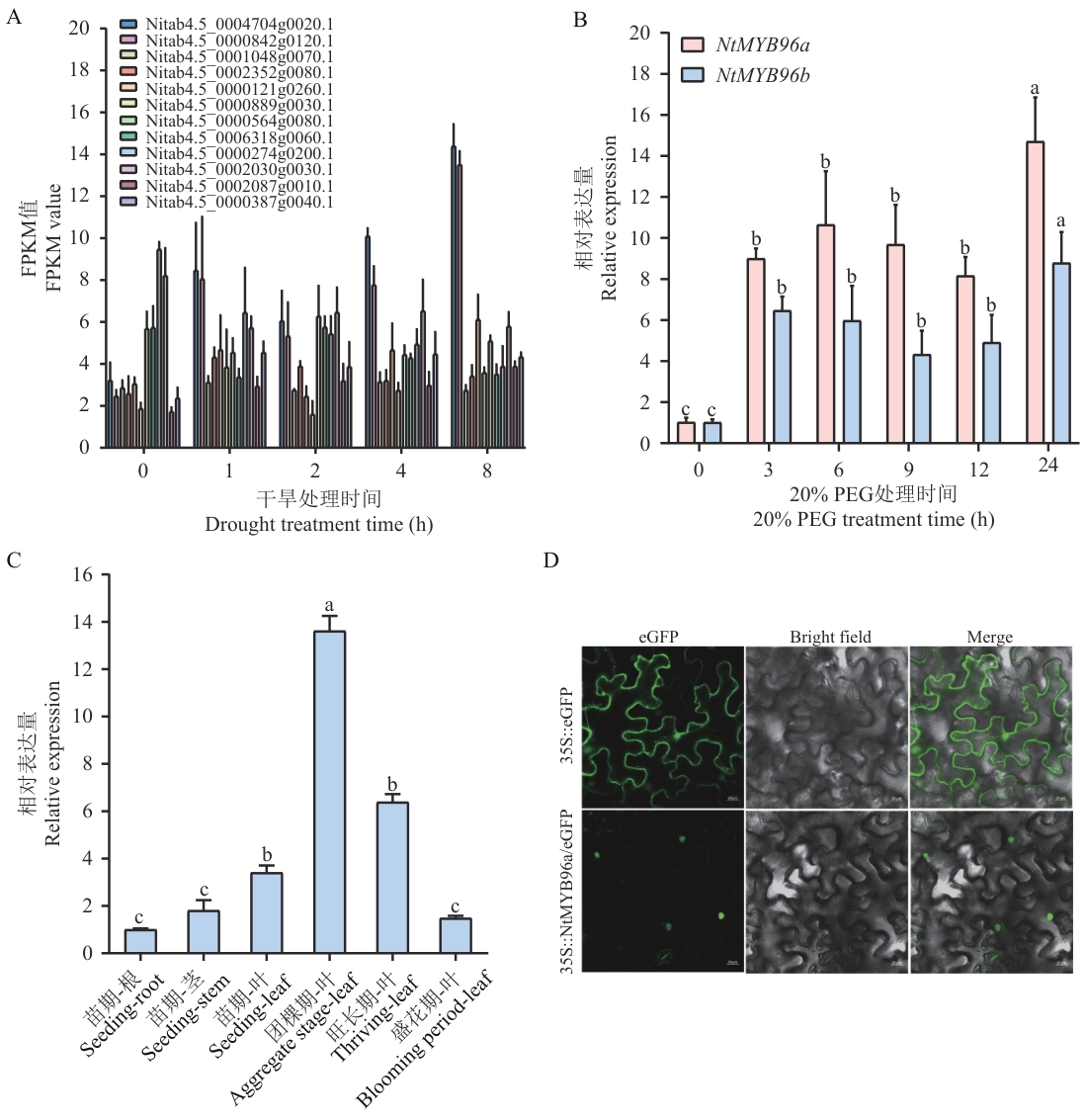
Fig. 3 Screening and expression characterization analysis of the NtMYB96a geneA: FPKM values of NtMYB96s genes from transcriptome data under drought stress. B: Expressions of NtMYB96a and NtMYB96bunder drought stress. C: Expression analysis of NtMYB96a in tissues of tobacco seedlings and in leaves at different developmental stages. D: Subcellular localization analysis of NtMYB96a. Different lowercase letters indicate statistically significant differences at P<0.05 (t test). All data are presented as mean ± SD from three biological replicates. The same below
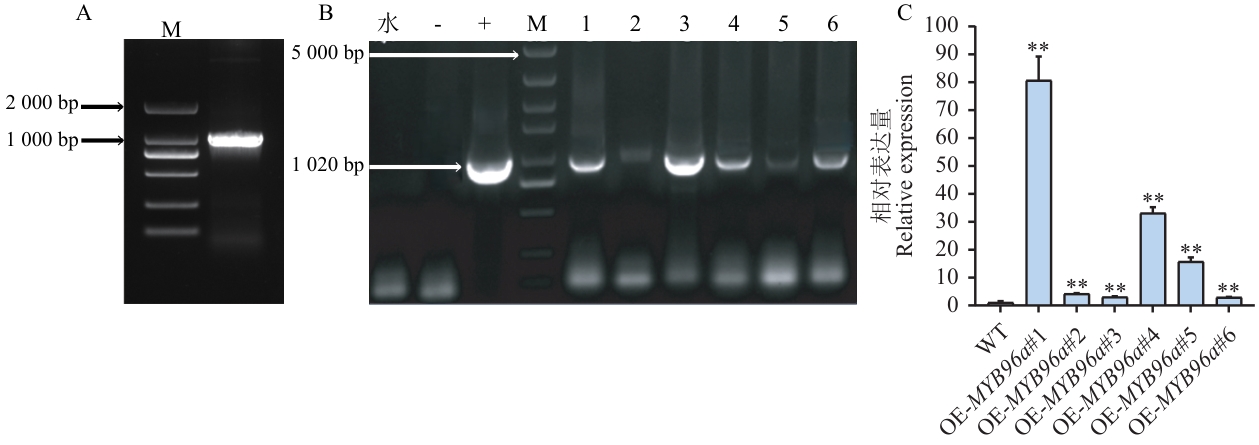
Fig. 4 Cloning and generation of overexpressing lines of NtMYB96aA: Amplification of the CDS region of NtMYB96a gene. B: Identification of positive transgenic tobacco plants overexpressing NtMYB96a (M: marker; -: negative control; +: positive control; 1-6: T0 generation positive plants). C: Expression analysis of NtMYB96a in overexpression plants. Asterisks indicate statistically significant differences compared with the wild-type (WT) plants. t test, three biological replicates, * P<0.05, ** P<0.01. The same below
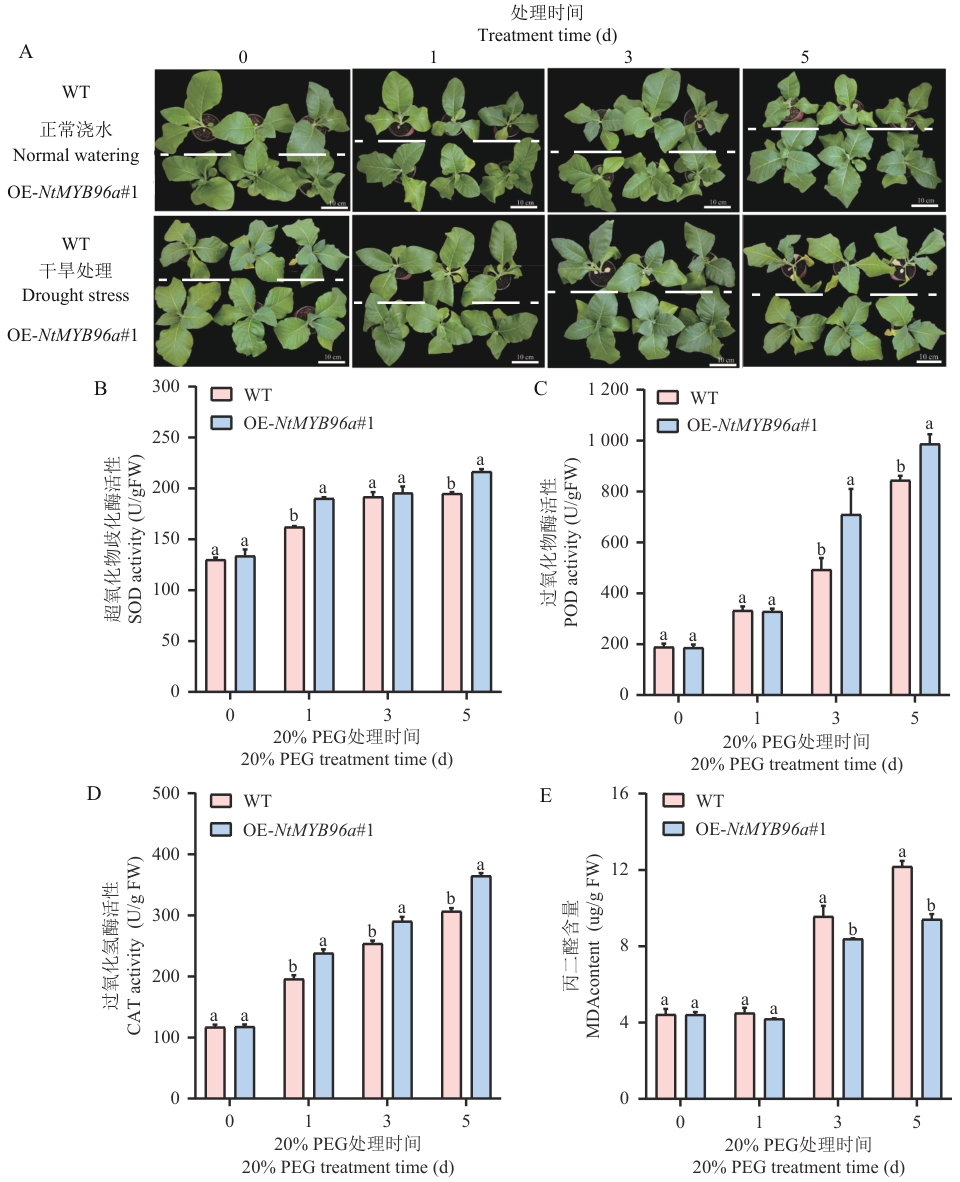
Fig. 5 Determination of seedling growth, antioxidant enzyme activity, and MDA content ofOE-NtMYB96a#1plants under drought stressA: Growing phenotype of plants under drought stress treatment (scale bar=10 cm). B-E: Analysis of antioxidant enzyme activity and MDA content under drought stress
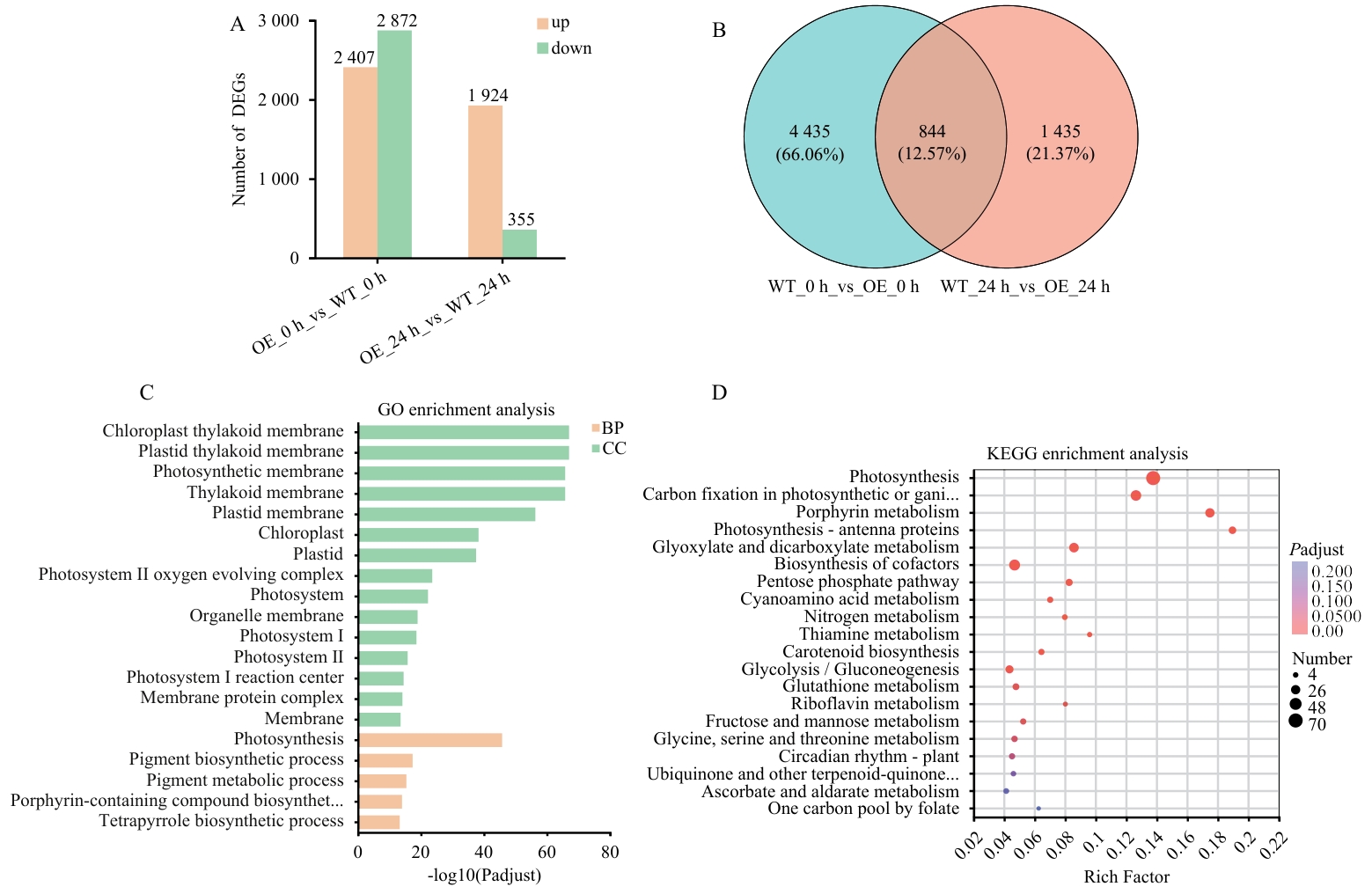
Fig. 6 Effects of NtMYB96a overexpression on the transcriptome of tobacco under drought stressA: Number of differentially expressed genes(DEGs). B: Venn diagram of DEGs. In panel A and B, the four groups are: WT under drought treatment at 0 h (WT_0 h), OE-NtMYB96a#1 under drought treatment at 0 h (OE_0 h), WT under drought treatment at 24 h (WT_24 h) , and OE-NtMYB96a#1 under drought treatment at 24 h (OE_24 h) . C: GO enrichment analysis of OE-NtMYB96a#1-specific DEGs. CC: Cellular component. BP: Biological process. D: KEGG enrichment analysis of DEGs unique to OE-NtMYB96a#1
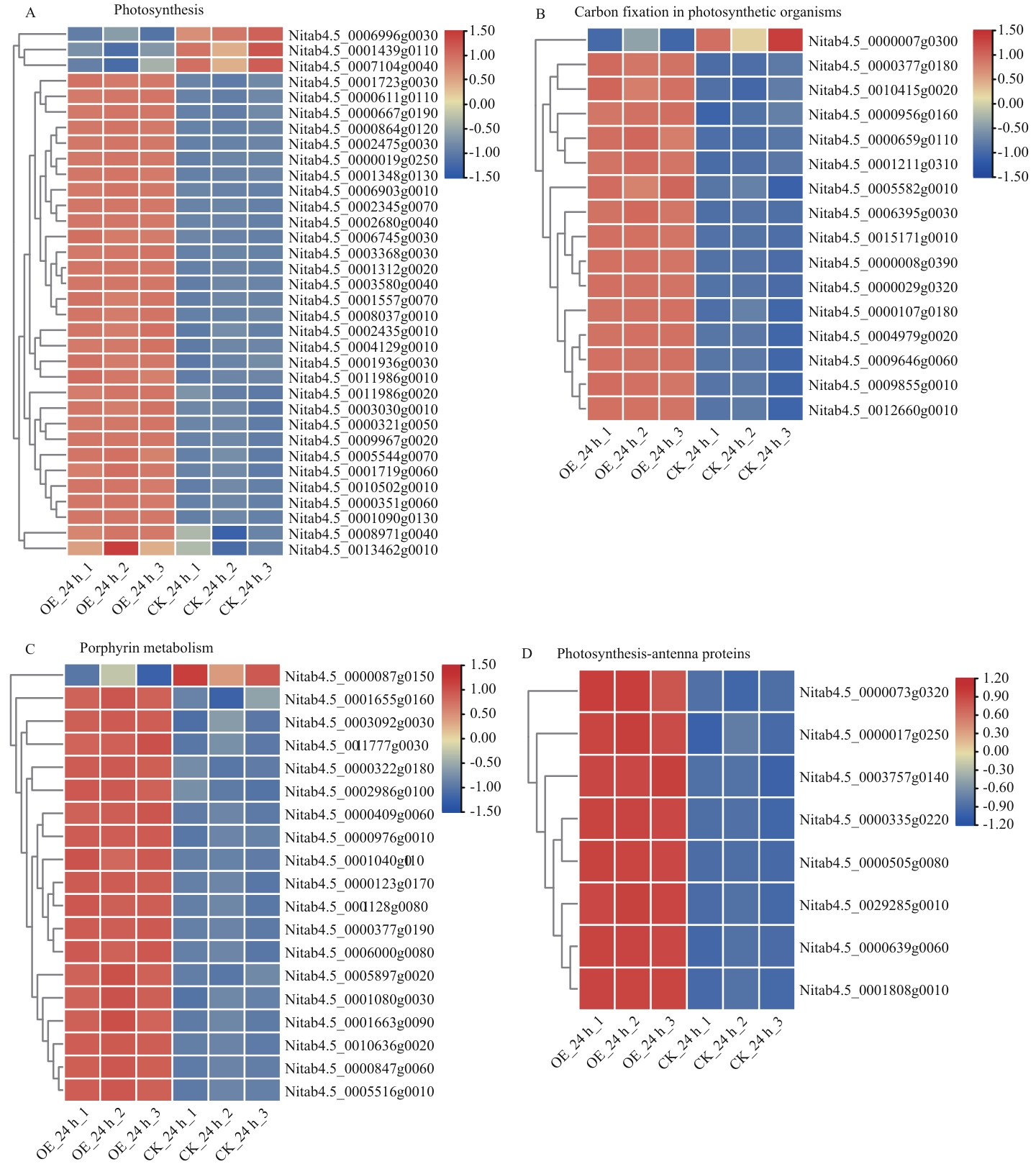
Fig. 7 Heatmap analysis of DEGsspecifically regulated by NtMYB96a overexpressionunder drought stressA: Photosynthesis. B: Carbon fixation of photosynthetic organisms. C: Porphyrin metabolism. D: Photosynthesis-antenna protein
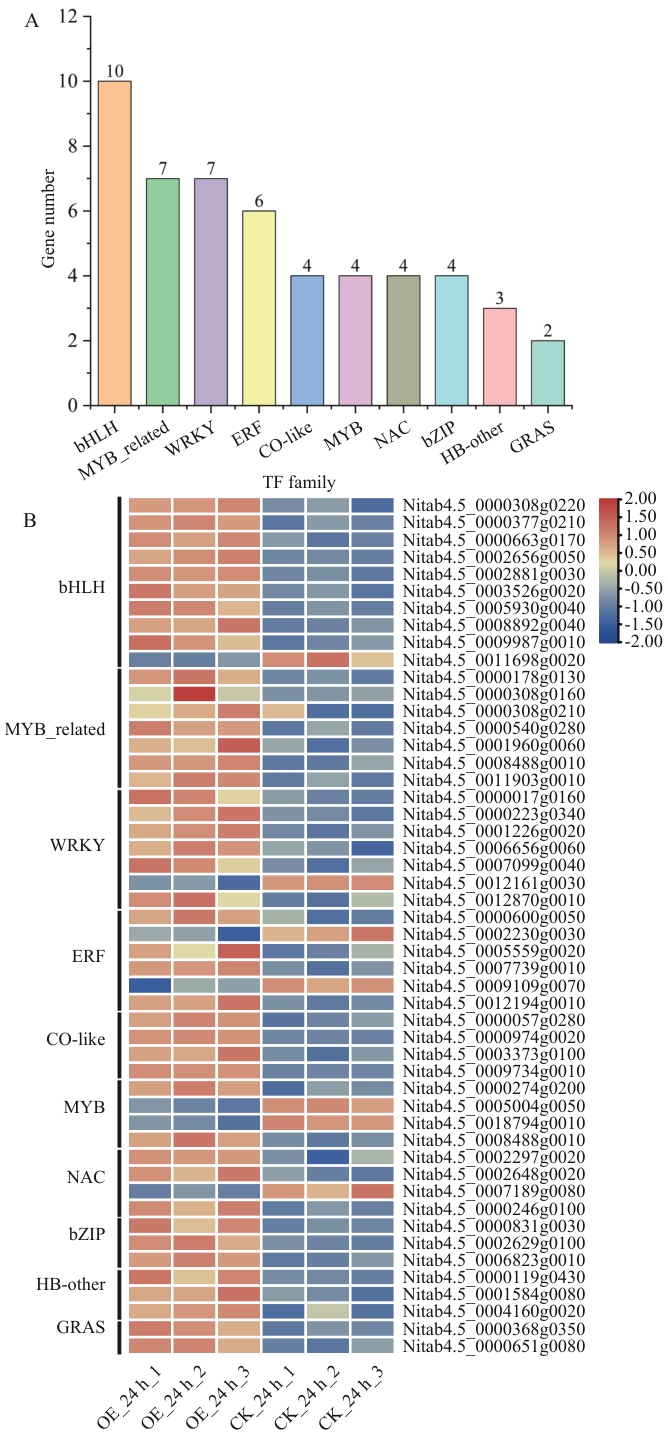
Fig. 8 Analysis of transcription factor families among DEGs specifically regulated by NtMYB96a overexpression under drought stressA: Statistics of transcription factor families. B: Expression analysis of transcription factor family members under drought stress
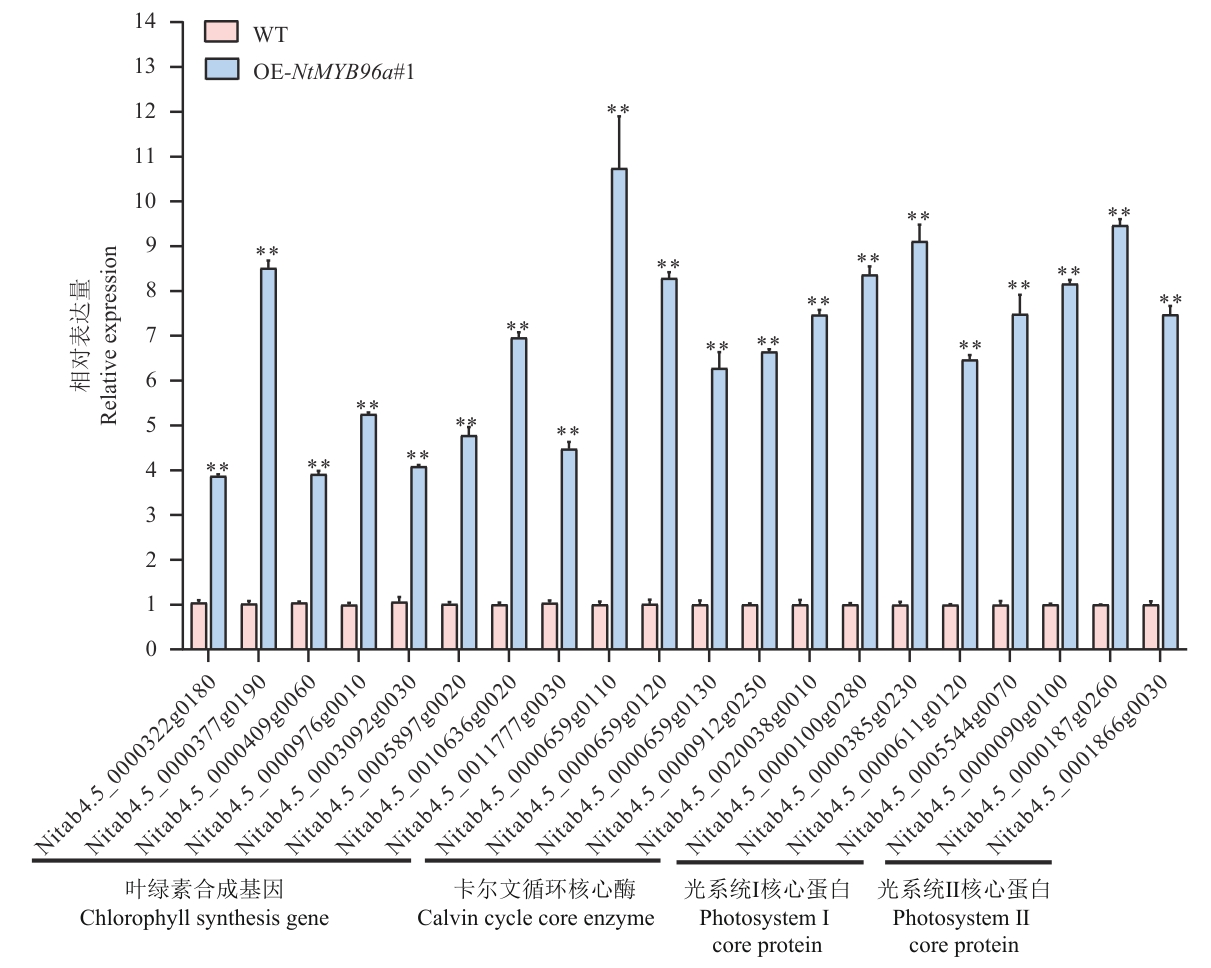
Fig. 9 Expressions of genes related to chlorophyll biosynthesis and photosynthesis in wild-type and NtMYB96a -overexpressing plants under drought stress
| [1] | 彭文婷, 李庆, 代明球. 作物抗旱关键调控因子的挖掘与利用 [J]. 科学通报, 2025, 70(19): 3149-3167. |
| Peng WT, Li Q, Dai MQ. Exploitation of key regulatory factors and breeding strategies for crop drought resistance [J]. Chin Sci Bull, 2025, 70(19): 3149-3167. | |
| [2] | 李佳, 刘涛, 马菊莲, 等. 烟草响应干旱胁迫与抗旱遗传育种研究进展 [J]. 江苏农业科学, 2023, 51(8): 34-43. |
| Li J, Liu T, Ma JL, et al. Research progress on drought stress response and drought resistance genetic breeding of tobacco [J]. Jiangsu Agric Sci, 2023, 51(8): 34-43. | |
| [3] | Zhang DW, Zhou HP, Zhang Y, et al. Diverse roles of MYB transcription factors in plants [J]. J Integr Plant Biol, 2025, 67(3): 539-562. |
| [4] | Liu YH, Shen Y, Liang M, et al. Identification of peanut AhMYB44 transcription factors and their multiple roles in drought stress responses [J]. Plants, 2022, 11(24): 3522. |
| [5] | Zhao HY, Zhao HQ, Hu YF, et al. Expression of the sweet potato MYB transcription factor IbMYB48 confers salt and drought tolerance in Arabidopsis [J]. Genes, 2022, 13(10): 1883. |
| [6] | Gao J, Zhao Y, Zhao ZK, et al. RRS1 shapes robust root system to enhance drought resistance in rice [J]. New Phytol, 2023, 238(3): 1146-1162. |
| [7] | Pil Joon Seo SBL. The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis [J]. Plant Cell, 2011, 23(3): 1138-1152. |
| [8] | 李霞, 张泽伟, 刘泽军, 等. 马铃薯转录因子StMYB96的克隆及功能研究 [J]. 生物技术通报, 2025, 41(7): 181-192. |
| Li X, Zhang ZW, Liu ZJ, et al. Cloning and functional study of transcription factor StMYB96 in potato [J]. Biotechnol Bull, 2025, 41(7): 181-192. | |
| [9] | He JJ, Li CZ, Hu N, et al. ECERIFERUM1-6A is required for the synthesis of cuticular wax alkanes and promotes drought tolerance in wheat [J]. Plant Physiol, 2022, 190(3): 1640-1657. |
| [10] | 赵先炎, 齐晨辉, 姜翰, 等. 苹果MpMYB96基因克隆和功能鉴定 [J]. 植物生理学报, 2019, 55(6): 783-792. |
| Zhao XY, Qi CH, Jiang H, et al. Cloning and functional identification of MpMYB96 gene in Malus pumila [J]. Plant Physiol J, 2019, 55(6): 783-792. | |
| [11] | 赵海英, 刘致远, 曹际玲. 纳米银对不同品种玉米幼苗生长和生理特性的影响 [J]. 热带农业科学, 2025, 45(7): 50-57. |
| Zhao HY, Liu ZY, Cao JL. Effects of silver nanoparticles on the growth and physiological characteristics of different varieties of maize seedlings [J]. Chin J Trop Agric, 2025, 45(7): 50-57. | |
| [12] | 孙美华, 孙慧贤, 赵晏俪, 等. 不同供氮水平对番茄果实心室数量的影响 [J]. 园艺学报, 2025, 52(10): 2737-2747. |
| Sun MH, Sun HX, Zhao YL, et al. Effects of different nitrogen concentration on the locule number in tomato fruit [J]. Acta Hortic Sin, 2025, 52(10): 2737-2747. | |
| [13] | Kim JB, Kim SH, Bae DH. The impacts of global warming on arid climate and drought features [J]. Theor Appl Climatol, 2023, 152(1): 693-708. |
| [14] | 王亚虹, 许自成, 高森, 等. 烟草干旱胁迫研究进展 [J]. 节水灌溉, 2016(12): 103-107, 111. |
| Wang YH, Xu ZC, Gao S, et al. Research progress of tobacco drought stress [J]. Water Sav Irrig, 2016(12): 103-107, 111. | |
| [15] | 金伊楠, 张豪洋, 张璐翔, 等. 烟草抗旱相关基因的研究进展 [J]. 烟草科技, 2020, 53(3): 18-26. |
| Jin YN, Zhang HY, Zhang LX, et al. Research progress of drought-resistance related genes in tobacco [J]. Tob Sci Technol, 2020, 53(3): 18-26. | |
| [16] | Wei QH, Zhang F, Sun FS, et al. A wheat MYB transcriptional repressor TaMyb1D regulates phenylpropanoid metabolism and enhances tolerance to drought and oxidative stresses in transgenic tobacco plants [J]. Plant Sci, 2017, 265: 112-123. |
| [17] | Liu ZY, Li JN, Li S, et al. The 1R-MYB transcription factor SlMYB1L modulates drought tolerance via an ABA-dependent pathway in tomato [J]. Plant Physiol Biochem, 2025, 222: 109721. |
| [18] | Yan ML, Li XX, Ji XY, et al. An R2R3-MYB transcription factor PdbMYB6 enhances drought tolerance by mediating reactive oxygen species scavenging, osmotic balance, and stomatal opening [J]. Plant Physiol Biochem, 2025, 220: 109536. |
| [19] | Zhang XB, Lei L, Lai JS, et al. Effects of drought stress and water recovery on physiological responses and gene expression in maize seedlings [J]. BMC Plant Biol, 2018, 18(1): 68. |
| [20] | Wang YX, Zhang MH, Li XY, et al. Overexpression of the wheat TaPsb28 gene enhances drought tolerance in transgenic Arabidopsis [J]. Int J Mol Sci, 2023, 24(6): 5226. |
| [21] | Wang LX, Wang BW, Du QZ, et al. Allelic variation in PtoPsbW associated with photosynthesis, growth, and wood properties in Populus tomentosa [J]. Mol Genet Genomics, 2017, 292(1): 77-91. |
| [22] | Cui L, Zheng FY, Zhang DD, et al. Tomato methionine sulfoxide reductase B2 functions in drought tolerance by promoting ROS scavenging and chlorophyll accumulation through interaction with Catalase 2 and RBCS3B [J]. Plant Sci, 2022, 318: 111206. |
| [23] | Liu EL, Xu LL, Luo ZQ, et al. Transcriptomic analysis reveals mechanisms for the different drought tolerance of sweet potatoes [J]. Front Plant Sci, 2023, 14: 1136709. |
| [24] | Karami S, Shiran B, Ravash R. Molecular investigation of how drought stress affects chlorophyll metabolism and photosynthesis in leaves of C3 and C4 plant species: a transcriptome meta-analysis [J]. Heliyon, 2025, 11(3): e42368. |
| [25] | Wei GQ, Zhang YT, Yang Y, et al. CfCHLM, from Cryptomeria fortunei, promotes chlorophyll synthesis and improves tolerance to abiotic stresses in transgenic Arabidopsis thaliana [J]. Forests, 2024, 15(4): 628. |
| [26] | Ye JJ, Lin XY, Yang ZX, et al. The light-harvesting chlorophyll a/b-binding proteins of photosystem Ⅱ family members are responsible for temperature sensitivity and leaf color phenotype in albino tea plant [J]. J Adv Res, 2024, 66: 87-104. |
| [27] | Cheng JQ, Wu TT, Zhou Y, et al. The alternative splicing of HvLHCA4.2 enhances drought tolerance in barley by regulating ROS scavenging and stomatal closure [J]. Int J Biol Macromol, 2025, 307: 142384. |
| [28] | Zhang HY, Wang YL, Song XY, et al. BcLhcb2.1, a Light-harvesting chlorophyll a/b-binding protein from Wucai, plays a positive regulatory role in the response to abiotic stress [J]. Sci Hortic, 2025, 339: 113759. |
| [29] | Zhao S, Gao HB, Luo JW, et al. Genome-wide analysis of the light-harvesting chlorophyll a/b-binding gene family in apple (Malus domestica) and functional characterization of MdLhcb4.3, which confers tolerance to drought and osmotic stress [J]. Plant Physiol Biochem, 2020, 154: 517-529. |
| [30] | Liang YF, Ma F, Li BY, et al. A bHLH transcription factor, SlbHLH96, promotes drought tolerance in tomato [J]. Hortic Res, 2022, 9: uhac198. |
| [31] | Zhao PC, Hou SL, Guo XF, et al. A MYB-related transcription factor from sheepgrass, LcMYB2, promotes seed germination and root growth under drought stress [J]. BMC Plant Biol, 2019, 19(1): 564. |
| [32] | Gulzar F, Fu JY, Zhu CY, et al. Maize WRKY transcription factor ZmWRKY79 positively regulates drought tolerance through elevating ABA biosynthesis [J]. Int J Mol Sci, 2021, 22(18): 10080. |
| [1] | QIN Zi-lu, SUN Hai-yan, CHEN Ying-nan. Research Progress in the Molecular Regulatory Mechanism of Plant Trichome Development [J]. Biotechnology Bulletin, 2026, 42(9): 1-13. |
| [2] | YIN Ya-long, ZHANG Ming-yang, WANG Jie-min, MIAO Xue-xue, CHEN Jin, WANG Wei-ping. Advances in Coordinated Tolerance Mechanisms to Abiotic Stresses in Rice [J]. Biotechnology Bulletin, 2026, 42(4): 26-37. |
| [3] | SU Yan-zhu, LI Da, ZHANG Ai-ai, LIU Yong-guang, ZHANG Xiu-rong, XUE Qi-qin. Identification and Expression Analysis of CAD Gene Family in Soybean(Glycine max (L.) Merr.) [J]. Biotechnology Bulletin, 2026, 42(4): 101-113. |
| [4] | REN Yun-er, WU Guo-qiang, CHENG Bin, WEI Ming. Genome-wide Identification of the BvATGs Genes Family in Sugar Beet (Beta vulgaris L.) and Analysis of Their Expression Pattern under Salt Stress [J]. Biotechnology Bulletin, 2026, 42(1): 184-197. |
| [5] | YANG Yue-qin, XING Ying, ZHONG Zi-he, TIAN Wei-jun, YANG Xue-qing, WANG Jian-xu. Expression and Functional Analysis of OsMATE34 in Rice under Mercury Stress [J]. Biotechnology Bulletin, 2026, 42(1): 86-94. |
| [6] | ZHANG Chi-hao, LIU Jin-nan, CHAO Yue-hui. Cloning and Functional Analysis of a bZIP Transcription Factor MtbZIP29 from Medicago truncatula [J]. Biotechnology Bulletin, 2026, 42(1): 241-250. |
| [7] | ZHANG Yue, DAI Yue-hua, ZHANG Ying-ying, LI Ao-hui, LI Chu-hui, XUE Jin-ai, QIN Hui-bin, CHEN Yan, NIE Meng-en, ZHANG Hai-ping. Cloning and Functional Analysis of the Soybean Enoyl-CoA Reductase ECR14 Gene [J]. Biotechnology Bulletin, 2026, 42(1): 95-104. |
| [8] | CHEN Jing-huan, FANG Guo-nan, ZHU Wen-hao, YE Guang-ji, SU Wang, HE Miao-miao, YANG Sheng-long, ZHOU Yun. Starch Characterization and Related Gene Expression Analysis of Potato Germplasm Resources [J]. Biotechnology Bulletin, 2026, 42(1): 170-183. |
| [9] | LI Shan, MA Deng-hui, MA Hong-yi, YAO Wen-kong, YIN Xiao. Identification and Expression Analysis of SKP1 Gene Family in Grapevine (Vitis vinifera L.) [J]. Biotechnology Bulletin, 2025, 41(9): 147-158. |
| [10] | GONG Hui-ling, XING Yu-jie, MA Jun-xian, CAI Xia, FENG Zai-ping. Identification of Laccase (LAC) Gene Family in Potato (Solanum tuberosum L.) and Its Expression Analysis under Salt Stresses [J]. Biotechnology Bulletin, 2025, 41(9): 82-93. |
| [11] | HUA Wen-ping, LIU Fei, HAO Jia-xin, CHEN Chen. Identification and Expression Patterns Analysis of ADH Gene Family in Salvia miltiorrhiza [J]. Biotechnology Bulletin, 2025, 41(8): 211-219. |
| [12] | ZHAI Ying, JI Jun-jie, CHEN Jiong-xin, YU Hai-wei, LI Shan-shan, ZHAO Yan, MA Tian-yi. Heterologous Overexpression of Soybean GmNF-YB24 Improves the Resistance of Transgenic Tobacco to Drought [J]. Biotechnology Bulletin, 2025, 41(8): 137-145. |
| [13] | REN Rui-bin, SI Er-jing, WAN Guang-you, WANG Jun-cheng, YAO Li-rong, ZHANG Hong, MA Xiao-le, LI Bao-chun, WANG Hua-jun, MENG Ya-xiong. Identification and Expression Analysis of GH17 Gene Family of Pyrenophora graminea [J]. Biotechnology Bulletin, 2025, 41(8): 146-154. |
| [14] | ZENG Dan, HUANG Yuan, WANG Jian, ZHANG Yan, LIU Qing-xia, GU Rong-hui, SUN Qing-wen, CHEN Hong-yu. Genome-wide Identification and Expression Analysis of bZIP Transcription Factor Family in Dendrobium officinale [J]. Biotechnology Bulletin, 2025, 41(8): 197-210. |
| [15] | CHENG Xue, FU Ying, CHAI Xiao-jiao, WANG Hong-yan, DENG Xin. Identification of LHC Gene Family in Setaria italica and Expression Analysis under Abiotic Stresses [J]. Biotechnology Bulletin, 2025, 41(8): 102-114. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||