








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (3): 121-129.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0471
Previous Articles Next Articles
FU Ya-li( ), PENG Wan-li, LIN Shuang-jun, DENG Zi-xin, LIANG Ru-bing(
), PENG Wan-li, LIN Shuang-jun, DENG Zi-xin, LIANG Ru-bing( )
)
Received:2021-04-11
Online:2022-03-26
Published:2022-04-06
Contact:
LIANG Ru-bing
E-mail:fuyali_1502@163.com;icelike@sjtu.edu.cn
FU Ya-li, PENG Wan-li, LIN Shuang-jun, DENG Zi-xin, LIANG Ru-bing. Gene Cloning and Enzymatic Properties of the Short Chain Dehydrogenase SDR-X1 from Pseudomonas citronellolis SJTE-3[J]. Biotechnology Bulletin, 2022, 38(3): 121-129.

Fig. 1 Multiple sequence alignment,advanced structural model and evolutionary analysis of SDR-X1 protein A:Multiple sequence alignment of short chain dehydrogenase from different microorganisms. Protein SDR-X1(WP_043267487.1)from P. citronellol SJTE-3,protein CAD85559.1 from N. europaea ATCC19718,protein WP_010595991.1 and WP_010595922.1 from R. ruber P14,protein WP_061563290.1 from P. citronellolis P3B5,protein WP_008027336.1 from P. putida B6-2,protein ABQ79984.1 from Pseudomonas putida F1,protein WP_012271104.1 from Pseudomonas putida GB-1,protein WP_014754112.1/ANI02794.1/ANI04816.1 from P. putida SJTE-1,protein WP_057091786.1 from C.testosteroni P19,and protein WP_010128053.1 from Sphingomonas KC8. The spiral marks the α-helix and the arrow marks the β-sheet. B:Having 4iqg.1.A as a model,and a three-dimensional model of protein obtained from homologous modeling of protein SDR-X1 with SWISSMODEL. C:Evolutionary analysis of the protein SDR-X1
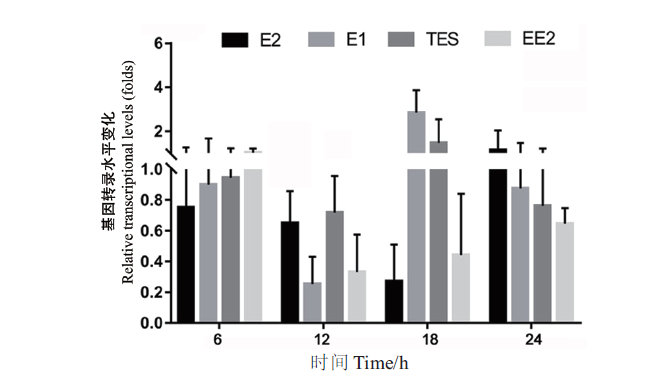
Fig. 2 Transcription levels of the gene sdr-x1 in strain SJTE-3 with different steroid hormones as carbon sources The carbon source is 10 μg/mL of steroid hormones(E1,E2,EE2 or TES)or 0.1% ethanol,and the transcription level of the gene sdr-x1 in ethanol as carbon source medium is set as 1.0. The results of fluorescence quantitative PCR amplification were processed by 2-∆∆Ct method. Three parallel experiments were set for each group and repeated three times,and based on which the mean and standard error were calculated

Fig. 3 Efficacy determination of recombinant strain BL21-SDR-X1 for transforming E2,purification of recombinant protein SDR-X1 and its transformation reaction to E2 A:The efficacy determination of recombinant strain BL21-SDR-X1 for transforming E2. The conversion efficiency of strain BL21-SDR-X1 to 10 μg/mL 17β-estradiol at 3,6,12,18 and 24 h is detected by HPLC. The control group is BL21(DE3)strain with plasmid pET28a. Three parallel experiments are set in each group and repeated for three times,and based on it the mean and standard error are calculated. B:Electrophoresis detection of affinity-purified recombinant protein SDR-X1. Lane 1:protein maker;lane 2:cell lysis solution before induction;lane 3:cell lysis solution after induction;lane 4:centrifugal precipitation of the crushed cells;lane 5:centrifugal supernatant of the crushed cells;lane 6-7:the solution after column loading;lane 8-9:the impurities;lane 10:protein eluent. C:Kinetic curve of recombinant protein SDR-X1 transforming E2. Using E2 as substrate,the effect of protein SDR-X1 on different concentration(0.0100,0.0125,0.0150,0.0200,0.0250,0.0300 0,0.0500 0,0.075,and 0.1 mmol/L)substrate is determined. Three parallel experiments are set for each group and repeated three times,and the mean and standard error are calculated
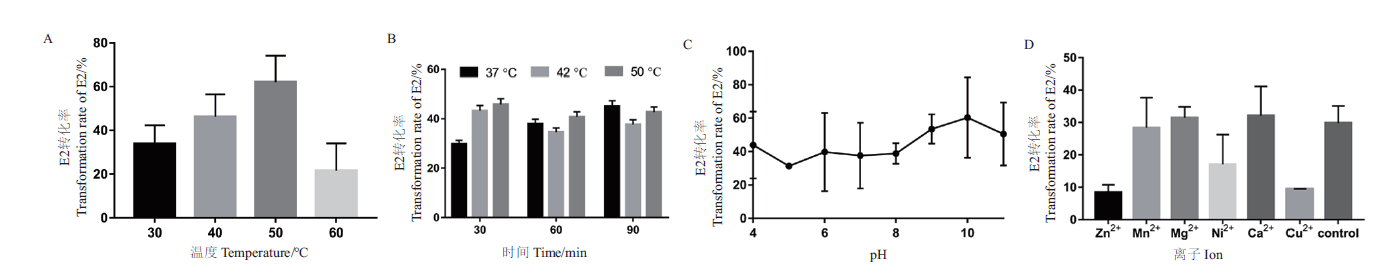
Fig. 4 Determination of the enzymatic properties of E2 transformed by recombinant enzyme SDR-X1 A:Determination of optimal reaction temperature. The conversion efficiency of recombinase SDR-X1 at different temperature(30,40,50,and 60℃)was detected with E2 as substrate. B:Temperature tolerance was measured. The conversion efficiency of recombinase SDR-X1 treated with different temperature(37,42,and 50℃)and time(30,60,and 90 min)was detected with E2 as substrate. C:Optimal reaction pH determination. The conversion efficiency of recombinase SDR-X1 at different pH value(pH4-11)was detected with E2 as substrate. D:Determination of influence of divalent metal ions. The conversion efficiency of recombinase SDR-X1 under the action of different metal ion(Zn2+,Mn2+,Mg2+,Ni2+,Ca2+,and Cu2+)at 1 mmol/L was determined. Three experiments were set in parallel for each group and repeated for three times. The mean value and standard error are calculated
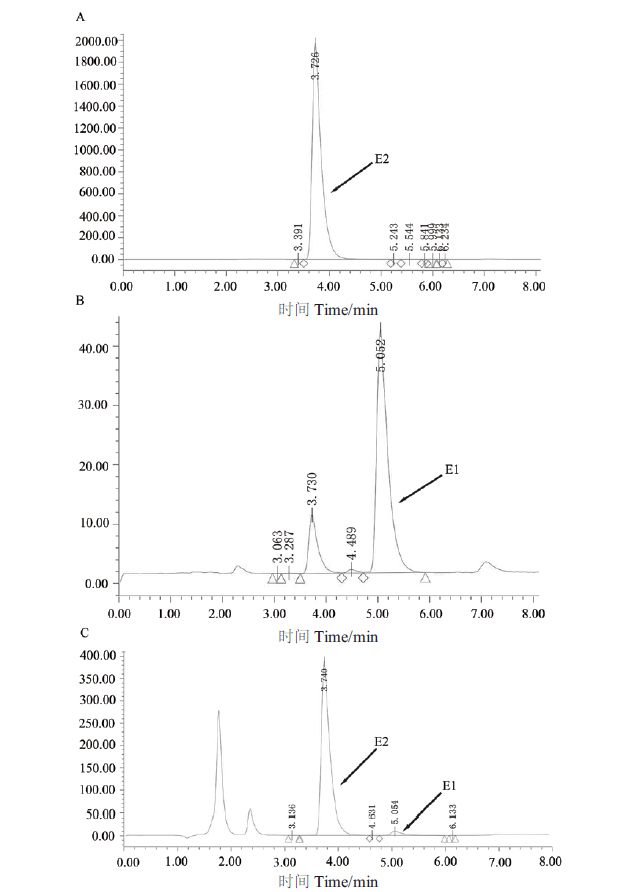
Fig. 5 HPLC analysis of reaction products of conversion of E2 by recombinase SDR-X1 A:The HPLC map of 50 μg/mL E2. B:The HPLC map of 50 μg/mL E1. C:The HPLC map of the products of E2 convered by recombinase SDR-X1. The reaction solution is 0.1 mmol/L E2,0.2 mmol/L NAD+ and 100 μg/mL protein SDR-X1 in a 200 μL reaction system at 42℃ for 15 min. Then adding 1/3 volume of acetonitrile to the reaction solution,swirling and mixing,50 μL sample is taken to detect degradation of E2 by HPLC
| [1] |
Wise A, O'Brien K, Woodruff T. Are oral contraceptives a significant contributor to the estrogenicity of drinking water?[J]. Environ Sci Technol, 2011, 45(1):51-60.
doi: 10.1021/es1014482 URL |
| [2] |
Kiyama R, Wada-Kiyama Y. Estrogenic endocrine disruptors:Molecular mechanisms of action[J]. Environ Int, 2015, 83:11-40.
doi: 10.1016/j.envint.2015.05.012 URL |
| [3] |
Chen YL, Wang CH, Yang FC, et al. Identification of Comamonas testosteroni as an androgen degrader in sewage[J]. Sci Rep, 2016, 6:35386.
doi: 10.1038/srep35386 URL |
| [4] |
Luine VN. Estradiol and cognitive function:past, present and future[J]. Horm Behav, 2014, 66(4):602-618.
doi: 10.1016/j.yhbeh.2014.08.011 URL |
| [5] |
Donova MV, Egorova OV. Microbial steroid transformations:current state and prospects[J]. Appl Microbiol Biotechnol, 2012, 94(6):1423-1447.
doi: 10.1007/s00253-012-4078-0 URL |
| [6] | Chen YL, Fu HY, Lee TH, et al. Estrogen degraders and estrogen degradation pathway identified in an activated sludge[J]. Appl Environ Microbiol, 2018, 84(10):e00001-e00018. |
| [7] |
Bai X, Acharya K. Removal of seven endocrine disrupting chemicals(EDCs)from municipal wastewater effluents by a freshwater green alga[J]. Environ Pollut, 2019, 247:534-540.
doi: 10.1016/j.envpol.2019.01.075 URL |
| [8] |
Xiong W, Yin C, Wang Y, et al. Characterization of an efficient estrogen-degrading bacterium Stenotrophomonas maltophilia SJTH1 in saline-, alkaline-, heavy metal-contained environments or solid soil and identification of four 17β-estradiol-oxidizing dehydrogenases[J]. J Hazard Mater, 2020, 385:121616.
doi: 10.1016/j.jhazmat.2019.121616 URL |
| [9] |
Wang P, Zheng D, Liang R. Isolation and characterization of an estrogen-degrading Pseudomonas putida strain SJTE-1[J]. 3 Biotech, 2019, 9(2):61.
doi: 10.1007/s13205-018-1537-z URL |
| [10] | Li SY, Liu J, Sun MX, et al. Isolation, characterization, and degradation performance of the 17β-estradiol-degrading bacterium Novosphingobium sp. E2S[J]. Int J Environ Res Public Heal, 2017, 14(2):115. |
| [11] |
Xiong WL, Yin C, Peng WL, et al. Characterization of an 17β-estradiol-degrading bacterium Stenotrophomonas maltophilia SJTL3 tolerant to adverse environmental factors[J]. Appl Microbiol Biotechnol, 2020, 104(3):1291-1305.
doi: 10.1007/s00253-019-10281-8 URL |
| [12] |
Mascotti ML, Palazzolo MA, Bisogno FR, et al. Biotransformation of dehydro-epi-androsterone by Aspergillus parasiticus:Metabolic evidences of BVMO activity[J]. Steroids, 2016, 109:44-49.
doi: 10.1016/j.steroids.2016.03.018 pmid: 27025973 |
| [13] | Liu WJ, Chen Q, He N, et al. Removal and biodegradation of 17β-estradiol and diethylstilbestrol by the freshwater microalgae Raphidocelis subcapitata[J]. Int J Environ Res Public Heal, 2018, 15(3):452. |
| [14] |
Wang PH, Yu CP, Lee TH, et al. Anoxic androgen degradation by the denitrifying bacterium Sterolibacterium denitrificans via the 2, 3-seco pathway[J]. Appl Environ Microbiol, 2014, 80(11):3442-3452.
doi: 10.1128/AEM.03880-13 URL |
| [15] |
Wang PP, Zheng DN, Peng WL, et al. Characterization of 17β-hydroxysteroid dehydrogenase and regulators involved in estrogen degradation in Pseudomonas putida SJTE-1[J]. Appl Microbiol Biotechnol, 2019, 103(5):2413-2425.
doi: 10.1007/s00253-018-9543-y URL |
| [16] |
Ye X, Wang H, Kan J, et al. A novel 17β-hydroxysteroid dehydrogenase in Rhodococcus sp. P14 for transforming 17β-estradiol to estrone[J]. Chem Biol Interact, 2017, 276:105-112.
doi: 10.1016/j.cbi.2017.06.010 URL |
| [17] | Liu C, Liu K, Zhao C, et al. The characterization of a short chain dehydrogenase/reductase(SDRx)in Comamonas testosteroni[J]. Toxicol Rep, 2020, 7:460-467. |
| [18] |
Yu YH, Liu CZ, Wang BX, et al. Characterization of 3, 17β-hydroxysteroid dehydrogenase in Comamonas testosteroni[J]. Chem Biol Interact, 2015, 234:221-228.
doi: 10.1016/j.cbi.2015.01.005 URL |
| [19] |
Pan T, Huang P, Xiong G, et al. Isolation and identification of a repressor TetR for 3, 17β-HSD expressional regulation in Comamonas testosteroni[J]. Chem Biol Interact, 2015, 234:205-212.
doi: 10.1016/j.cbi.2014.12.034 URL |
| [20] |
Kavanagh KL, Jörnvall H, Persson B, et al. Medium- and short-chain dehydrogenase/reductase gene and protein families[J]. Cell Mol Life Sci, 2008, 65(24):3895-3906.
doi: 10.1007/s00018-008-8588-y pmid: 19011750 |
| [21] |
Gräff M, Buchholz PCF, Stockinger P, et al. The Short-chain Dehydrogenase/Reductase Engineering Database(SDRED):a classification and analysis system for a highly diverse enzyme family[J]. Proteins, 2019, 87(6):443-451.
doi: 10.1002/prot.v87.6 URL |
| [22] | Zheng DN, Wang XL, Wang PP, et al. Genome sequence of Pseudomonas citronellolis SJTE-3, an estrogen-and polycyclic aromatic hydrocarbon-degrading bacterium[J]. Genome Announc, 2016, 4(6):e01373-16. |
| [23] |
Hall BG. Building phylogenetic trees from molecular data with MEGA[J]. Mol Biol Evol, 2013, 30(5):1229-1235.
doi: 10.1093/molbev/mst012 URL |
| [24] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method[J]. Methods, 2001, 25(4):402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [25] |
Xu J, Zhang L, Hou J, et al. iTRAQ-based quantitative proteomic analysis of the global response to 17β-estradiol in estrogen-degradation strain Pseudomonas putida SJTE-1[J]. Sci Rep, 2017, 7:41682.
doi: 10.1038/srep41682 URL |
| [26] |
Kallberg Y, Oppermann U, Persson B. Classification of the short-chain dehydrogenase/reductase superfamily using hidden Markov models[J]. Febs J, 2010, 277(10):2375-2386.
doi: 10.1111/j.1742-4658.2010.07656.x pmid: 20423462 |
| [27] | Mythen SM, Devendran S, Méndez-García C, et al. Targeted synjournal and characterization of a gene cluster encoding NAD(P)H-dependent 3α-, 3β-, and 12α-hydroxysteroid dehydrogenases from Eggerthella CAG:298, a gut metagenomic sequence[J]. Appl Environ Microbiol, 2018, 84(7):e02475-17. |
| [28] |
Zhao BH, Sun Q, Chen J, et al. 17 beta-estradiol biodegradation by anaerobic granular sludge:Effect of iron sources[J]. Sci Rep, 2020, 10:7777.
doi: 10.1038/s41598-020-64557-5 URL |
| [1] | WANG Hao, TANG Lu-xin, MA Hong-fei, QIAN Kun, SI Jing, CUI Bao-kai. Immobilization of Laccase from Trametes orientalis and Its Application for Decolorization of Multifarious Dyes [J]. Biotechnology Bulletin, 2021, 37(11): 142-157. |
| [2] | WU Min, TANG Jie, HU Qiong, LEI Dan, ZHANG Qing. Effects of Surfactants on Deltamethrin Degradation by Acinetobacter junii LH-1-1 [J]. Biotechnology Bulletin, 2021, 37(1): 215-222. |
| [3] | YUE Li-xiao, LI Deng-yun, ZHANG Jing-jing, TONG Lei. Isolation and Application Potential Exploration of a Diuron-degrading Bacterium [J]. Biotechnology Bulletin, 2020, 36(6): 110-119. |
| [4] | CHEN Rui, QU Jia, SUN Xiao-yu, DENG Yuan, MEN Xin, ZHAO Ling-xia, SHEN Wei-rong. Isolation and Identification of Penicillium oxalicum SSCL-5 Degrading Cypermethrin and Its Biodegradation [J]. Biotechnology Bulletin, 2020, 36(6): 120-127. |
| [5] | REN Lei, LIU Bin, LIN Zhong, ZHEN Zhen, LIU Yue-lian, HU Han-qiao, YAN Yan-chun. Isolation of a p-Nitrophenol-Degrading Bacterium and Investigation of Its Degrading Mechanism [J]. Biotechnology Bulletin, 2019, 35(9): 184-193. |
| [6] | GUO Ya-nan, ZHANG Xin-yu, XU Meng, WANG Ji-hua. Screening,Identification of Naphthalene Degrading Bacteria at Low Temperature and Optimization of Their Degradation Conditions [J]. Biotechnology Bulletin, 2019, 35(7): 100-107. |
| [7] | LIU Ya-lan, DUAN Meng-jie, LIN Xiao-shan, ZHANG Yi. Identification and Characterization of Bacteria Degrading Polyvinyl Alcohol [J]. Biotechnology Bulletin, 2019, 35(6): 91-98. |
| [8] | ZHANG Shan-shan, YAO Xiao-long, WANG Ke, YOU Ya. Degradation of Ethyl Formate by Aeromonas salmonicida subsp.salmonicida [J]. Biotechnology Bulletin, 2019, 35(5): 109-117. |
| [9] | TIAN Jing, XU Xiao-lin, KANG Yan-shun, TANG Wei-hua, LIU Si-qi. Screening and Characteristics of a Broad Spectrum Fungus Degrading Polycyclic-aromatic Hydrocarbons:Aspergillus flavus AD-X-1 [J]. Biotechnology Bulletin, 2018, 34(8): 115-122. |
| [10] | SUN Gui-ting ,CHEN Hong-yun ,ZHAO Ling-chao ,HU Xiao-ke ,CHEN Ying. Isolation,Identification and Characterization of a PCBs-degrading Bacterium [J]. Biotechnology Bulletin, 2018, 34(6): 141-148. |
| [11] | CHEN Rui, SUN Xiao-yu, DENG Yuan, LU Peng-peng, ZHAO Ling-xia, QU Jia, SHEN Wei-rong. Biological Characteristic and Biodegradation Capacity of Ochrobactrum tritici WNP-3 for Carbendazim [J]. Biotechnology Bulletin, 2018, 34(5): 187-194. |
| [12] | WANG Jia-yi FAN, Shuang-hu, REN Chao, WANG Jun-huan, YANG Ting, JIA Yang, LI Xian-jun, YAN Yan-chun. Identification of Newly Isolated Xanthobacter sp. and Its Degradability to Phthalic Acid Esters [J]. Biotechnology Bulletin, 2018, 34(10): 157-164. |
| [13] | TIAN Lin, ZHANG Xun. Study on the Changes of Soil Microbial Community Structure in Farmland by Estrone Stress [J]. Biotechnology Bulletin, 2017, 33(6): 230-236. |
| [14] | FENG Yan-mei, FAN Xing-hui, ZHAN Hui, TENG Shi-yu, YANG Fang, CHEN Shao-hua. Research Progress on Ecotoxicity and Microbial Degradation of Strobilurin Fungicides [J]. Biotechnology Bulletin, 2017, 33(10): 52-58. |
| [15] | LIU Shu-yan, WANG Fang, WANG Jian-yu, LIN Rong-shan. Isolation,Identification and Degrading Properties of Phlorizin-Degrading Fungi [J]. Biotechnology Bulletin, 2017, 33(10): 143-147. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||