








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (5): 159-168.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1071
Previous Articles Next Articles
LI Yu-hang1,2( ), WANG Xing-ping1,2(
), WANG Xing-ping1,2( ), YANG Jian1,2, LUORENG Zhuo-ma1,2, REN Qian-qian1,2, WEI Da-wei1,2, MA Yun1,2
), YANG Jian1,2, LUORENG Zhuo-ma1,2, REN Qian-qian1,2, WEI Da-wei1,2, MA Yun1,2
Received:2021-08-20
Online:2022-05-26
Published:2022-06-10
Contact:
WANG Xing-ping
E-mail:1429692080@qq.com;wxp@nxu.edu.cn
LI Yu-hang, WANG Xing-ping, YANG Jian, LUORENG Zhuo-ma, REN Qian-qian, WEI Da-wei, MA Yun. Expression and Functional Analysis of miR-665 in Bovine Mammary Epithelial Cell Inflammation[J]. Biotechnology Bulletin, 2022, 38(5): 159-168.
| 基因名称 Gene name | 引物序列 Prime sequence(5'-3') | 产物大小 Product length/bp |
|---|---|---|
| miR-665 | RT:GTCGTATCCAGTGCAGGGTCCGAGG TATTCGCACTGGATACGACAGGGGCCT | 64 |
| F:GTCATGGAACCAGTAGGCCG | ||
| R:CAGTGCAGGGTCCGAGGTAT | ||
| IL-1β | F:CAACCGTACCTGAACCC | 193 |
| R:GACACCACCTGCCTGAA | ||
| IL-8 | F:ACACATTCCACACCTTTCCAC | 149 |
| R:ACCTTCTGCACCCACTTTTC | ||
| GADPH | F:GGCATCGTGGAGGGACTTATG | 186 |
| R:GCCAGTGAGCTTCCCGTTGAG | ||
| RPS18 | F:GTGGTGTTGAGGAAAGCAGACA | 79 |
| R:TGATCACACGTTCCACCTCATC |
Table 1 Reverse transcription and qPCR primers of miR-665
| 基因名称 Gene name | 引物序列 Prime sequence(5'-3') | 产物大小 Product length/bp |
|---|---|---|
| miR-665 | RT:GTCGTATCCAGTGCAGGGTCCGAGG TATTCGCACTGGATACGACAGGGGCCT | 64 |
| F:GTCATGGAACCAGTAGGCCG | ||
| R:CAGTGCAGGGTCCGAGGTAT | ||
| IL-1β | F:CAACCGTACCTGAACCC | 193 |
| R:GACACCACCTGCCTGAA | ||
| IL-8 | F:ACACATTCCACACCTTTCCAC | 149 |
| R:ACCTTCTGCACCCACTTTTC | ||
| GADPH | F:GGCATCGTGGAGGGACTTATG | 186 |
| R:GCCAGTGAGCTTCCCGTTGAG | ||
| RPS18 | F:GTGGTGTTGAGGAAAGCAGACA | 79 |
| R:TGATCACACGTTCCACCTCATC |
| 基因名称 Gene name | 引物序列 Prime sequence(5'-3') | 产物大小 Product length/bp |
|---|---|---|
| MAPK14 | F:ATCTTGGATTTCGGACTGGC | 264 |
| R:ATGGCTTGGCATCCTGTTA | ||
| MAP3K2 | F:CGGAAATACACCCGTCAGA | 324 |
| R:AGCCATCGCTTCAAACTCG | ||
| SMAD2 | F:GAAGGCAGACGGTAACAAGC | 114 |
| R:TTGAGCAACGCACTGAAGG | ||
| SP1 | F:CGGTGGGCAGTATGTTGT | 237 |
| R:CCTCCACTTCCTCGGTTT |
Table 2 qPCR primers for potential key target genes of miR-665
| 基因名称 Gene name | 引物序列 Prime sequence(5'-3') | 产物大小 Product length/bp |
|---|---|---|
| MAPK14 | F:ATCTTGGATTTCGGACTGGC | 264 |
| R:ATGGCTTGGCATCCTGTTA | ||
| MAP3K2 | F:CGGAAATACACCCGTCAGA | 324 |
| R:AGCCATCGCTTCAAACTCG | ||
| SMAD2 | F:GAAGGCAGACGGTAACAAGC | 114 |
| R:TTGAGCAACGCACTGAAGG | ||
| SP1 | F:CGGTGGGCAGTATGTTGT | 237 |
| R:CCTCCACTTCCTCGGTTT |
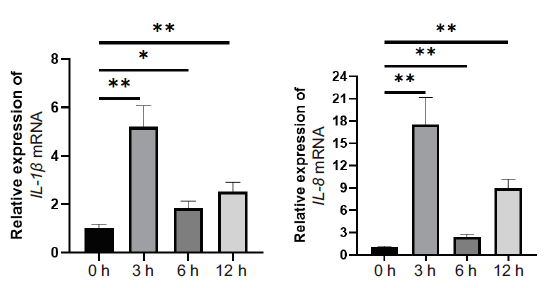
Fig.1 Expression of major inflammatory factors in the LPS-induced inflammation of mammary epithelial cells * refers to P < 0.05;** refers to P < 0.01,the same below
| 基因名称Gene name | 牛Bos taurus | 人Homo sapiens | 小鼠Mus musculus | ||||
|---|---|---|---|---|---|---|---|
| GenBank ID | GenBank ID | 同源性Homology/% | GenBank ID | 同源性Homology/% | |||
| MAPK14 | NM_001102174.1 | NM_001315.3 | 84.34 | NM_011951.3 | 84.88 | ||
| MAP3K2 | XM_002685232.6 | NM_006609.5 | 85.91 | NM_011946.3 | 85.14 | ||
| SMAD2 | NM_001046218.1 | NM_005901.6 | 93.96 | NM_010754.5 | 88.70 | ||
| SP1 | NM_001078027.1 | NM_138473.3 | 91.03 | NM_013672.2 | 88.99 |
Table 3 Homology analysis of related genes in the bovine,human and mouse
| 基因名称Gene name | 牛Bos taurus | 人Homo sapiens | 小鼠Mus musculus | ||||
|---|---|---|---|---|---|---|---|
| GenBank ID | GenBank ID | 同源性Homology/% | GenBank ID | 同源性Homology/% | |||
| MAPK14 | NM_001102174.1 | NM_001315.3 | 84.34 | NM_011951.3 | 84.88 | ||
| MAP3K2 | XM_002685232.6 | NM_006609.5 | 85.91 | NM_011946.3 | 85.14 | ||
| SMAD2 | NM_001046218.1 | NM_005901.6 | 93.96 | NM_010754.5 | 88.70 | ||
| SP1 | NM_001078027.1 | NM_138473.3 | 91.03 | NM_013672.2 | 88.99 |
| [1] |
He W, Ma S, Lei L, et al. Prevalence, etiology, and economic impact of clinical mastitis on large dairy farms in China[J]. Vet Microbiol, 2020, 242:108570.
doi: 10.1016/j.vetmic.2019.108570 URL |
| [2] |
Royster E, Wagner S. Treatment of mastitis in cattle[J]. Vet Clin North Am Food Anim Pract, 2015, 31(1):17-46, v.
doi: 10.1016/j.cvfa.2014.11.010 URL |
| [3] |
Gorji AE, Roudbari Z, Sadeghi B, et al. Transcriptomic analysis on the promoter regions discover gene networks involving mastitis in cattle[J]. Microb Pathog, 2019, 137:103801.
doi: 10.1016/j.micpath.2019.103801 URL |
| [4] |
Asselstine V, Miglior F, Suárez-Vega A, et al. Genetic mechanisms regulating the host response during mastitis[J]. J Dairy Sci, 2019, 102(10):9043-9059.
doi: S0022-0302(19)30701-5 pmid: 31421890 |
| [5] |
Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, et al. An overview of microRNAs:Biology, functions, therapeutics, and analysis methods[J]. J Cell Physiol, 2019, 234(5):5451-5465.
doi: 10.1002/jcp.27486 pmid: 30471116 |
| [6] |
Correia de Sousa M, Gjorgjieva M, Dolicka D, et al. Deciphering miRNAs’ action through miRNA Editing[J]. Int J Mol Sci, 2019, 20(24):6249.
doi: 10.3390/ijms20246249 URL |
| [7] |
Rupaimoole R, Slack FJ. MicroRNA therapeutics:towards a new era for the management of cancer and other diseases[J]. Nat Rev Drug Discov, 2017, 16(3):203-222.
doi: 10.1038/nrd.2016.246 pmid: 28209991 |
| [8] |
Luoreng ZM, Wang XP, Mei CG, et al. Comparison of microRNA profiles between bovine mammary glands infected with Staphylococcus aureus and Escherichia coli[J]. Int J Biol Sci, 2018, 14(1):87-99.
doi: 10.7150/ijbs.22498 URL |
| [9] |
Li R, Zhang CL, Liao XX, et al. Transcriptome MicroRNA profiling of bovine mammary glands infected with Staphylococcus aureus[J]. Int J Mol Sci, 2015, 16(12):4997-5013.
doi: 10.3390/ijms16034997 URL |
| [10] |
Luoreng ZM, Yang J, Wang XP, et al. Expression profiling of microRNA from peripheral blood of dairy cows in response to Staphylococcus aureus-infected mastitis[J]. Front Vet Sci, 2021, 8:691196.
doi: 10.3389/fvets.2021.691196 URL |
| [11] |
Luoreng ZM, Wang XP, Mei CG, et al. Expression profiling of peripheral blood miRNA using RNAseq technology in dairy cows with Escherichia coli-induced mastitis[J]. Sci Rep, 2018, 8(1):12693.
doi: 10.1038/s41598-018-30518-2 URL |
| [12] |
Wang XP, Luoreng ZM, Zan LS, et al. Bovine miR-146a regulates inflammatory cytokines of bovine mammary epithelial cells via targeting the TRAF6 gene[J]. J Dairy Sci, 2017, 100(9):7648-7658.
doi: 10.3168/jds.2017-12630 URL |
| [13] |
Cai MC, Fan WQ, Li XY, et al. The regulation of Staphylococcus aureus-induced inflammatory responses in bovine mammary epithelial cells[J]. Front Vet Sci, 2021, 8:683886.
doi: 10.3389/fvets.2021.683886 URL |
| [14] |
Li M, Zhang S, Qiu Y, et al. Upregulation of miR-665 promotes apoptosis and colitis in inflammatory bowel disease by repressing the endoplasmic Reticulum stress components XBP1 and ORMDL3[J]. Cell Death Dis, 2017, 8(3):e2699.
doi: 10.1038/cddis.2017.76 URL |
| [15] | Liu S, Li XM, Yuan JB, et al. MiR-665 inhibits inflammatory response in microglia following spinal cord injury by targeting TREM2[J]. Eur Rev Med Pharmacol Sci, 2021, 25(1):65-70. |
| [16] | Guo Q, Lin Y, Hu J. Inhibition of miR-665-3p enhances autophagy and alleviates inflammation in Fusarium solani-induced keratitis[J]. Invest Ophthalmol Vis Sci, 2021, 62(1):24. |
| [17] |
Pareek R, Wellnitz O, Van Dorp R, et al. Immunorelevant gene expression in LPS-challenged bovine mammary epithelial cells[J]. J Appl Genet, 2005, 46(2):171-177.
pmid: 15876684 |
| [18] |
Strandberg Y, Gray C, Vuocolo T, et al. Lipopolysaccharide and lipoteichoic acid induce different innate immune responses in bovine mammary epithelial cells[J]. Cytokine, 2005, 31(1):72-86.
pmid: 15882946 |
| [19] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T))Method[J]. Methods, 2001, 25(4):402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [20] |
Sharun K, Dhama K, Tiwari R, et al. Advances in therapeutic and managemental approaches of bovine mastitis:a comprehensive review[J]. Vet Q, 2021, 41(1):107-136.
doi: 10.1080/01652176.2021.1882713 pmid: 33509059 |
| [21] |
Dalanezi FM, Joaquim SF, Guimarães FF, et al. Influence of pathogens causing clinical mastitis on reproductive variables of dairy cows[J]. J Dairy Sci, 2020, 103(4):3648-3655.
doi: S0022-0302(20)30114-4 pmid: 32089296 |
| [22] |
Xu H, Zhang T, Hu X, et al. Exosomal miR-193b-5p as a regulator of LPS-induced inflammation in dairy cow mammary epithelial cells[J]. In Vitro Cell Dev Biol Anim, 2021, 57(7):695-703.
doi: 10.1007/s11626-021-00596-0 URL |
| [23] |
Chen Z, Xu X, Tan T, et al. MicroRNA-145 regulates immune cytokines via targeting FSCN1 in Staphylococcus aureus-induced mastitis in dairy cows[J]. Reprod Domest Anim, 2019, 54(6):882-891.
doi: 10.1111/rda.13438 pmid: 30974481 |
| [24] |
Zhang M, Wang S, Yi A, et al. microRNA-665 is down-regulated in gastric cancer and inhibits proliferation, invasion, and EMT by targeting PPP2R2A[J]. Cell Biochem Funct, 2020, 38(4):409-418.
doi: 10.1002/cbf.3485 URL |
| [25] | Liu J, Jiang Y, Wan Y, et al. MicroRNA-665 suppresses the growth and migration of ovarian cancer cells by targeting HOXA10[J]. Mol Med Rep, 2018, 18(3):2661-2668. |
| [26] |
Zhao XG, Hu JY, Tang J, et al. miR-665 expression predicts poor survival and promotes tumor metastasis by targeting NR4A3 in breast cancer[J]. Cell Death Dis, 2019, 10(7):479.
doi: 10.1038/s41419-019-1705-z URL |
| [27] |
Damasceno LEA, Prado DS, Veras FP, et al. PKM2 promotes Th17 cell differentiation and autoimmune inflammation by fine-tuning STAT3 activation[J]. J Exp Med, 2020, 217(10):e20190613. DOI: 10.1084/jem,20190613.
doi: 10.1084/jem,20190613 URL |
| [28] |
Bougarne N, Weyers B, Desmet SJ, et al. Molecular actions of PPARα in lipid metabolism and inflammation[J]. Endocr Rev, 2018, 39(5):760-802.
doi: 10.1210/er.2018-00064 pmid: 30020428 |
| [29] |
Rahman A, Henry KM, Herman KD, et al. Inhibition of ErbB kinase signalling promotes resolution of neutrophilic inflammation[J]. Elife, 2019, 8:e50990.
doi: 10.7554/eLife.50990 URL |
| [30] |
Clarke DC, Lauffenburger DA. Multi-pathway network analysis of mammalian epithelial cell responses in inflammatory environments[J]. Biochem Soc Trans, 2012, 40(1):133-138.
doi: 10.1042/BST20110633 URL |
| [31] |
Madkour MM, Anbar HS, El-Gamal MI. Current status and future prospects of p38α/MAPK14 kinase and its inhibitors[J]. Eur J Med Chem, 2021, 213:113216.
doi: 10.1016/j.ejmech.2021.113216 URL |
| [32] |
Pan W, Wei N, Xu W, et al. MicroRNA-124 alleviates the lung injury in mice with septic shock through inhibiting the activation of the MAPK signaling pathway by downregulating MAPK14[J]. Int Immunopharmacol, 2019, 76:105835.
doi: 10.1016/j.intimp.2019.105835 URL |
| [33] |
Zhang L, Han B, Liu H, et al. Circular RNA circACSL1 aggravated myocardial inflammation and myocardial injury by sponging miR-8055 and regulating MAPK14 expression[J]. Cell Death Dis, 2021, 12(5):487.
doi: 10.1038/s41419-021-03777-7 pmid: 33986259 |
| [34] |
Schmidt C, Peng B, Li Z, et al. Mechanisms of proinflammatory cytokine-induced biphasic NF-kappaB activation[J]. Mol Cell, 2003, 12(5):1287-1300.
doi: 10.1016/S1097-2765(03)00390-3 URL |
| [35] | Wang AD, Dai LF, Yang L, et al. Upregulation of miR-335 reduces myocardial injury following myocardial infarction via targeting MAP3K2[J]. Eur Rev Med Pharmacol Sci, 2021, 25(1):344-352. |
| [36] |
Wu N, Chen D, Sun H, et al. MAP3K2 augments Th1 cell differentiation via IL-18 to promote T cell-mediated colitis[J]. Sci China Life Sci, 2021, 64(3):389-403.
doi: 10.1007/s11427-020-1720-9 URL |
| [37] |
Martinez GJ, Zhang Z, Reynolds JM, et al. Smad2 positively regulates the generation of Th17 cells[J]. J Biol Chem, 2010, 285(38):29039-29043.
doi: 10.1074/jbc.C110.155820 pmid: 20667820 |
| [38] |
Kao YH, Chen PH, Wu TY, et al. Lipopolysaccharides induce Smad2 phosphorylation through PI3K/Akt and MAPK cascades in HSC-T6 hepatic stellate cells[J]. Life Sci, 2017, 184:37-46.
doi: 10.1016/j.lfs.2017.07.004 URL |
| [39] | Zhou YX, Han WW, Song DD, et al. Effect of miR-10a on Sepsis-induced liver injury in rats through TGF-β1/Smad signaling pathway[J]. Eur Rev Med Pharmacol Sci, 2020, 24(2):862-869. |
| [40] |
Jia L, Sun P, Gao H, et al. Mangiferin attenuates bleomycin-induced pulmonary fibrosis in mice through inhibiting TLR4/p65 and TGF-β1/Smad2/3 pathway[J]. J Pharm Pharmacol, 2019, 71(6):1017-1028.
doi: 10.1111/jphp.13077 pmid: 30847938 |
| [41] |
Chen L, Yang T, Lu DW, et al. Central role of dysregulation of TGF-β/Smad in CKD progression and potential targets of its treatment[J]. Biomed Pharmacother, 2018, 101:670-681.
doi: S0753-3322(17)36823-3 pmid: 29518614 |
| [42] |
Yang Q, Ren GL, Wei B, et al. Conditional knockout of TGF-βRII /Smad2 signals protects against acute renal injury by alleviating cell necroptosis, apoptosis and inflammation[J]. Theranostics, 2019, 9(26):8277-8293.
doi: 10.7150/thno.35686 pmid: 31754396 |
| [43] |
Cai F, Chen L, Sun Y, et al. MiR-539 inhibits the malignant behavior of breast cancer cells by targeting SP1[J]. Biochem Cell Biol, 2020, 98(3):426-433.
doi: 10.1139/bcb-2019-0111 URL |
| [44] |
Peng XB, Wu MH, Liu WX, et al. miR-502-5p inhibits the proliferation, migration and invasion of gastric cancer cells by targeting SP1[J]. Oncol Lett, 2020, 20(3):2757-2762.
doi: 10.3892/ol.2020.11808 URL |
| [45] |
Fu J, Li T, Jiang X, et al. MicroRNA-199-3p targets Sp1 transcription factor to regulate proliferation and epithelial to mesenchymal transition of human lung cancer cells[J]. 3 Biotech, 2021, 11(7):352.
doi: 10.1007/s13205-021-02881-x URL |
| [46] |
Wang R, Yang Y, Wang H, et al. MiR-29c protects against inflammation and apoptosis in Parkinson’s disease model in vivo and in vitro by targeting SP1[J]. Clin Exp Pharmacol Physiol, 2020, 47(3):372-382.
doi: 10.1111/1440-1681.13212 URL |
| [47] |
Lee WR, Kim KH, An HJ, et al. Effects of chimeric decoy oligodeoxynucleotide in the regulation of transcription factors NF-κB and Sp1 in an animal model of atherosclerosis[J]. Basic Clin Pharmacol Toxicol, 2013, 112(4):236-243.
doi: 10.1111/bcpt.12029 URL |
| [48] |
Zheng H, Dong X, Liu N, et al. Regulation and mechanism of mouse miR-130a/b in metabolism-related inflammation[J]. Int J Biochem Cell Biol, 2016, 74:72-83.
doi: 10.1016/j.biocel.2016.02.021 URL |
| [1] | WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(4): 246-258. |
| [2] | PING Huai-lei, GUO Xue, YU Xiao, SONG Jing, DU Chun, WANG Juan, ZHANG Huai-bi. Cloning and Expression of PdANS in Paeonia delavayi and Correlation with Anthocyanin Content [J]. Biotechnology Bulletin, 2023, 39(3): 206-217. |
| [3] | LI Yan-xia, WANG Jin-peng, FENG Fen, BAO Bin-wu, DONG Yi-wen, WANG Xing-ping, LUORENG Zhuo-ma. Effects of Escherichia coli Dairy Cow Mastitis on the Expressions of Milk-producing Trait Related Genes [J]. Biotechnology Bulletin, 2023, 39(2): 274-282. |
| [4] | GUO Zhi-hao, JIN Ze-xin, LIU Qi, GAO Li. Bioinformatics Analysis, Subcellular Localization and Toxicity Verification of Effector g11335 in Tilletia contraversa Kühn [J]. Biotechnology Bulletin, 2022, 38(8): 110-117. |
| [5] | YU Qiu-lin, MA Jing-yi, ZHAO Pan, SUN Peng-fang, HE Yu-mei, LIU Shi-biao, GUO Hui-hong. Cloning and Functional Analysis of Gynostemma pentaphyllum GpMIR156a and GpMIR166b [J]. Biotechnology Bulletin, 2022, 38(7): 186-193. |
| [6] | CHEN Jia-min, LIU Yong-jie, MA Jin-xiu, LI Dan, GONG Jie, ZHAO Chang-ping, GENG Hong-wei, GAO Shi-qing. Expression Pattern Analysis of Histone Methyltransferase Under Drought Stress in Hybrid Wheat [J]. Biotechnology Bulletin, 2022, 38(7): 51-61. |
| [7] | LIU Jing-jing, LIU Xiao-rui, LI Lin, WANG Ying, YANG Hai-yuan, DAI Yi-fan. Establishment of Porcine Fetal Fibroblasts with OXTR-knockout Using CRISPR/Cas9 [J]. Biotechnology Bulletin, 2022, 38(6): 272-278. |
| [8] | WANG Nan, ZHANG Rui, PAN Yang-yang, HE Hong-hong, WANG Jing-lei, CUI Yan, YU Si-jiu. Cloning of Bos grunniens TGF-β1 Gene and Its Expression in Major Organs of Female Reproductive System [J]. Biotechnology Bulletin, 2022, 38(6): 279-290. |
| [9] | LI Yang, ZHANG Xiao-tian, PIAO Jing-zi, ZHOU Ru-jun, LI Zi-bo, GUAN Hai-wen. Cloning and Bioinformatics Analysis of Blue-light Receptor EaWC 1 Gene in Elsinoë arachidis [J]. Biotechnology Bulletin, 2022, 38(5): 93-99. |
| [10] | ZHANG Lin, WEI Zhen-zhen, SONG Cheng-wei, GUO Li-li, GUO Qi, HOU Xiao-gai, WANG Hua-fang. Cloning and Expression Analysis of PoFD Gene from Paeonia ostii ‘Fengdan’ [J]. Biotechnology Bulletin, 2022, 38(11): 104-111. |
| [11] | ZHENG Qing-bo, YE Na, ZHANG Xiao-lan, BAO Peng-jia, WANG Fu-bin, REN Wen-wen, LIAO Yue-jiao, YAN Ping, PAN He-ping. Identification of Hair Follicle Cell Subsets and Bioinformatics Analysis of Characteristic Genes in Tianzhu White Yak During Catagen [J]. Biotechnology Bulletin, 2022, 38(10): 262-272. |
| [12] | FAN Ya-peng, RUI Cun, ZHANG Yue-xin, CHEN Xiu-gui, LU Xu-ke, WANG Shuai, ZHANG Hong, XU Nan, WANG Jing, CHEN Chao, YE Wu-wei. Cloning,Expression and Preliminary Bioinformatics Analysis of the Alkaline Tolerant Gene GhZAT12 in Gossypium hirsutum [J]. Biotechnology Bulletin, 2021, 37(8): 121-130. |
| [13] | DU Zhen-wei, ZHU Shuai-peng, MA Xiang-fei, LI Dong-hua, SUN Gui-rong. Cloning,Expression and Bioinformatics Analysis of the CDS Region of Chicken CEBPA Gene [J]. Biotechnology Bulletin, 2021, 37(8): 203-212. |
| [14] | HAO Xiang-yang, LIU Fan, WU Huan, WANG Bin, SUN Xue-li, XIANG Lei-lei, WANG Tian-chi, LAI Zhong-xiong, CHENG Chun-zhen. Cloning and Expression Analysis of GjPAL Genes in Gerbera jamesonni [J]. Biotechnology Bulletin, 2021, 37(6): 13-23. |
| [15] | WANG Jin-peng, LUORENG Zhuo-ma, WANG Xing-ping, YANG Jian, JIA Li, MA Yun, WEI Da-wei. Research Progress in Treatment and Anti-inflammatory Molecular Mechanism of Cow Mastitis [J]. Biotechnology Bulletin, 2021, 37(12): 212-219. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||